An extended consensus motif enhances the specificity of substrate modification by SUMO
- PMID: 17036045
- PMCID: PMC1630412
- DOI: 10.1038/sj.emboj.7601383
An extended consensus motif enhances the specificity of substrate modification by SUMO
Abstract
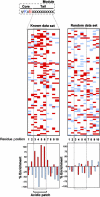
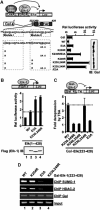
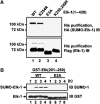
Protein modification by SUMO conjugation is an important regulatory event. Sumoylation usually takes place on a lysine residue embedded in the core consensus motif psiKxE. However, this motif confers limited specificity on the sumoylation process. Here, we have probed the roles of clusters of acidic residues located downstream from the core SUMO modification sites in proteins such as the transcription factor Elk-1. We demonstrate that these are functionally important in SUMO-dependent transcriptional repression of Elk-1 transcriptional activity. Mechanistically, the acidic residues are important in enhancing the efficiency of Elk-1 sumoylation by Ubc9. Similar mechanisms operate in other transcription factors and phosphorylation sites can functionally substitute for acidic residues. Thus, an extended sumoylation motif, termed the NDSM (negatively charged amino acid-dependent sumoylation motif), helps define functional SUMO targets. We demonstrate that this extended motif can be used to correctly predict novel targets for SUMO modification.
Figures

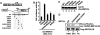


Similar articles
-
Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response.EMBO J. 2013 Mar 20;32(6):791-804. doi: 10.1038/emboj.2013.5. Epub 2013 Feb 8. EMBO J. 2013. PMID: 23395904 Free PMC article.
-
SUMO: getting it on.Biochem Soc Trans. 2007 Dec;35(Pt 6):1409-13. doi: 10.1042/BST0351409. Biochem Soc Trans. 2007. PMID: 18031233 Review.
-
Molecular mechanism of K65 acetylation-induced attenuation of Ubc9 and the NDSM interaction.Sci Rep. 2017 Dec 12;7(1):17391. doi: 10.1038/s41598-017-17465-0. Sci Rep. 2017. PMID: 29234076 Free PMC article.
-
In vitro modification of human centromere protein CENP-C fragments by small ubiquitin-like modifier (SUMO) protein: definitive identification of the modification sites by tandem mass spectrometry analysis of the isopeptides.J Biol Chem. 2004 Sep 17;279(38):39653-62. doi: 10.1074/jbc.M405637200. Epub 2004 Jul 22. J Biol Chem. 2004. PMID: 15272016
-
SUMO Ubc9 enzyme as a viral target.IUBMB Life. 2014 Jan;66(1):27-33. doi: 10.1002/iub.1240. Epub 2014 Jan 6. IUBMB Life. 2014. PMID: 24395713 Review.
Cited by
-
The Key Regulator for Language and Speech Development, FOXP2, is a Novel Substrate for SUMOylation.J Cell Biochem. 2016 Feb;117(2):426-38. doi: 10.1002/jcb.25288. J Cell Biochem. 2016. PMID: 26212494 Free PMC article.
-
Small ubiquitin-like modifier (SUMO) modification mediates function of the inhibitory domains of developmental regulators FOXC1 and FOXC2.J Biol Chem. 2012 May 25;287(22):18318-29. doi: 10.1074/jbc.M112.339424. Epub 2012 Apr 5. J Biol Chem. 2012. PMID: 22493429 Free PMC article.
-
Synthetic derivatives of the SUMO consensus sequence provide a basis for improved substrate recognition.Bioorg Med Chem Lett. 2015;25(10):2146-51. doi: 10.1016/j.bmcl.2015.03.069. Epub 2015 Mar 31. Bioorg Med Chem Lett. 2015. PMID: 25881829 Free PMC article.
-
Modification and movement: Phosphorylation and SUMOylation regulate endocytosis of GluK2-containing kainate receptors.Commun Integr Biol. 2012 Mar 1;5(2):223-6. doi: 10.4161/cib.19195. Commun Integr Biol. 2012. PMID: 22808340 Free PMC article.
-
Starting and stopping SUMOylation. What regulates the regulator?Chromosoma. 2013 Dec;122(6):451-63. doi: 10.1007/s00412-013-0422-0. Epub 2013 Jun 28. Chromosoma. 2013. PMID: 23812602 Review.
References
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 - PubMed
-
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

