Profile of histone lysine methylation across transcribed mammalian chromatin
- PMID: 17030614
- PMCID: PMC1698537
- DOI: 10.1128/MCB.01529-06
Profile of histone lysine methylation across transcribed mammalian chromatin
Abstract
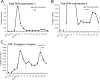
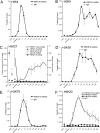
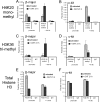
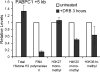
Complex patterns of histone lysine methylation encode distinct functions within chromatin. We previously reported that trimethylation of lysine 9 of histone H3 (H3K9) occurs at both silent heterochromatin and at the transcribed regions of active mammalian genes, suggesting that the extent of histone lysine methylation involved in mammalian gene activation is not completely defined. To identify additional sites of histone methylation that respond to mammalian gene activity, we describe here a comparative assessment of all six known positions of histone lysine methylation and relate them to gene transcription. Using several model loci, we observed high trimethylation of H3K4, H3K9, H3K36, and H3K79 in the transcribed region, consistent with previous findings. We identify H4K20 monomethylation, a modification previously linked with repression, as a mark of transcription elongation in mammalian cells. In contrast, H3K27 monomethylation, a modification enriched at pericentromeric heterochromatin, was observed broadly distributed throughout all euchromatic sites analyzed, with selective depletion in the vicinity of the transcription start sites at active genes. Together, these results underscore that similar to other described methyl-lysine modifications, H4K20 and H3K27 monomethylation are versatile and dynamic with respect to gene activity, suggesting the existence of novel site-specific methyltransferases and demethylases coupled to the transcription cycle.
Figures


Similar articles
-
Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin.Mol Cell. 2005 Aug 5;19(3):381-91. doi: 10.1016/j.molcel.2005.06.011. Mol Cell. 2005. PMID: 16061184
-
Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes.Mol Cell Biol. 2007 Dec;27(24):8466-79. doi: 10.1128/MCB.00993-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923682 Free PMC article.
-
Partitioning and plasticity of repressive histone methylation states in mammalian chromatin.Mol Cell. 2003 Dec;12(6):1577-89. doi: 10.1016/s1097-2765(03)00477-5. Mol Cell. 2003. PMID: 14690609
-
The indexing potential of histone lysine methylation.Novartis Found Symp. 2004;259:22-37; discussion 37-47, 163-9. Novartis Found Symp. 2004. PMID: 15171245 Review.
-
Biological function and regulation of histone and non-histone lysine methylation in response to DNA damage.Acta Biochim Biophys Sin (Shanghai). 2016 Jul;48(7):603-16. doi: 10.1093/abbs/gmw050. Epub 2016 May 23. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 27217472 Review.
Cited by
-
DOT1L-mediated H3K79 methylation in chromatin is dispensable for Wnt pathway-specific and other intestinal epithelial functions.Mol Cell Biol. 2013 May;33(9):1735-45. doi: 10.1128/MCB.01463-12. Epub 2013 Feb 19. Mol Cell Biol. 2013. PMID: 23428873 Free PMC article.
-
Ageing-Related Changes to H3K4me3, H3K27ac, and H3K27me3 in Purified Mouse Neurons.Cells. 2024 Aug 21;13(16):1393. doi: 10.3390/cells13161393. Cells. 2024. PMID: 39195281 Free PMC article.
-
The Ino80 complex prevents invasion of euchromatin into silent chromatin.Genes Dev. 2015 Feb 15;29(4):350-5. doi: 10.1101/gad.256255.114. Genes Dev. 2015. PMID: 25691465 Free PMC article.
-
Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions.Nucleic Acids Res. 2015 Feb 18;43(3):1869-82. doi: 10.1093/nar/gkv013. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605796 Free PMC article.
-
Chromatin measurements reveal contributions of synthesis and decay to steady-state mRNA levels.Mol Syst Biol. 2012 Jul 17;8:593. doi: 10.1038/msb.2012.23. Mol Syst Biol. 2012. PMID: 22806141 Free PMC article.
References
-
- Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280:17732-17736. - PubMed
-
- Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. - PubMed
-
- Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. - PubMed
-
- Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315-326. - PubMed
-
- Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
