Heat-shock promoters: targets for evolution by P transposable elements in Drosophila
- PMID: 17029562
- PMCID: PMC1592238
- DOI: 10.1371/journal.pgen.0020165
Heat-shock promoters: targets for evolution by P transposable elements in Drosophila
Abstract
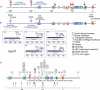
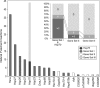
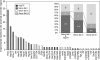
Transposable elements are potent agents of genomic change during evolution, but require access to chromatin for insertion-and not all genes provide equivalent access. To test whether the regulatory features of heat-shock genes render their proximal promoters especially susceptible to the insertion of transposable elements in nature, we conducted an unbiased screen of the proximal promoters of 18 heat-shock genes in 48 natural populations of Drosophila. More than 200 distinctive transposable elements had inserted into these promoters; greater than 96% are P elements. By contrast, few or no P element insertions segregate in natural populations in a "negative control" set of proximal promoters lacking the distinctive regulatory features of heat-shock genes. P element transpositions into these same genes during laboratory mutagenesis recapitulate these findings. The natural P element insertions cluster in specific sites in the promoters, with up to eight populations exhibiting P element insertions at the same position; laboratory insertions are into similar sites. By contrast, a "positive control" set of promoters resembling heat-shock promoters in regulatory features harbors few P element insertions in nature, but many insertions after experimental transposition in the laboratory. We conclude that the distinctive regulatory features that typify heat-shock genes (in Drosophila) are especially prone to mutagenesis via P elements in nature. Thus in nature, P elements create significant and distinctive variation in heat-shock genes, upon which evolutionary processes may act.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




Similar articles
-
Contrasting patterns of transposable element insertions in Drosophila heat-shock promoters.PLoS One. 2009 Dec 29;4(12):e8486. doi: 10.1371/journal.pone.0008486. PLoS One. 2009. PMID: 20041194 Free PMC article.
-
Abundant, diverse, and consequential P elements segregate in promoters of small heat-shock genes in Drosophila populations.J Evol Biol. 2007 Sep;20(5):2056-66. doi: 10.1111/j.1420-9101.2007.01348.x. J Evol Biol. 2007. PMID: 17714322
-
Remarkable site specificity of local transposition into the Hsp70 promoter of Drosophila melanogaster.Genetics. 2006 Jun;173(2):809-20. doi: 10.1534/genetics.105.053959. Epub 2006 Apr 2. Genetics. 2006. PMID: 16582443 Free PMC article.
-
Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster.Bioessays. 1994 Apr;16(4):269-75. doi: 10.1002/bies.950160410. Bioessays. 1994. PMID: 8031304 Review.
-
Gene disruptions using P transposable elements: an integral component of the Drosophila genome project.Proc Natl Acad Sci U S A. 1995 Nov 21;92(24):10824-30. doi: 10.1073/pnas.92.24.10824. Proc Natl Acad Sci U S A. 1995. PMID: 7479892 Free PMC article. Review.
Cited by
-
Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution.PLoS Genet. 2007 Jan 12;3(1):e10. doi: 10.1371/journal.pgen.0030010. Epub 2006 Dec 6. PLoS Genet. 2007. PMID: 17222062 Free PMC article.
-
A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen.Elife. 2021 Sep 16;10:e69249. doi: 10.7554/eLife.69249. Elife. 2021. PMID: 34528512 Free PMC article.
-
Divergence of Drosophila melanogaster repeatomes in response to a sharp microclimate contrast in Evolution Canyon, Israel.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10630-5. doi: 10.1073/pnas.1410372111. Epub 2014 Jul 8. Proc Natl Acad Sci U S A. 2014. PMID: 25006263 Free PMC article.
-
Key issues in achieving an integrative perspective on stress.J Biosci. 2007 Apr;32(3):433-40. doi: 10.1007/s12038-007-0042-z. J Biosci. 2007. PMID: 17536162
-
Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome.Genome Biol. 2006;7(11):R112. doi: 10.1186/gb-2006-7-11-r112. Genome Biol. 2006. PMID: 17134480 Free PMC article.
References
-
- Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Craig NL. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. - PubMed
-
- Engels WR. P elements in Drosophila . In: Saedler H, Gierl A, editors. Transposable elements. Berlin: Springer-Verlag; 1996. pp. 103–123.
-
- Castro JP, Carareto CMA. Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica. 2004;121:107–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

