Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex
- PMID: 17005651
- PMCID: PMC1676300
- DOI: 10.1128/JVI.01565-06
Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex
Abstract
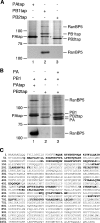
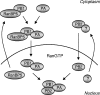
The influenza A virus RNA-dependent RNA polymerase is a heterotrimeric complex of polymerase basic protein 1 (PB1), PB2, and polymerase acidic protein (PA) subunits. It performs transcription and replication of the viral RNA genome in the nucleus of infected cells. We have identified a nuclear import factor, Ran binding protein 5 (RanBP5), also known as karyopherin beta3, importin beta3, or importin 5, as an interactor of the PB1 subunit. RanBP5 interacted with either PB1 alone or with a PB1-PA dimer but not with a PB1-PB2 dimer or the trimeric complex. The interaction between RanBP5 and PB1-PA was disrupted by RanGTP in vitro, allowing PB2 to bind to the PB1-PA dimer to form a functional trimeric RNA polymerase complex. We propose a model in which RanBP5 acts as an import factor for the newly synthesized polymerase by targeting the PB1-PA dimer to the nucleus. In agreement with this model, small interfering RNA (siRNA)-mediated knock-down of RanBP5 inhibited the nuclear accumulation of the PB1-PA dimer. Moreover, siRNA knock-down of RanBP5 resulted in the delayed accumulation of viral RNAs in infected cells, confirming that RanBP5 plays a biological role during the influenza virus life cycle.
Figures
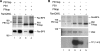

Similar articles
-
Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5.J Gen Virol. 2011 Aug;92(Pt 8):1859-1869. doi: 10.1099/vir.0.032813-0. Epub 2011 May 11. J Gen Virol. 2011. PMID: 21562121
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
-
Structural insights into Influenza A virus RNA polymerase PB1 binding to nuclear import host factor RanBP5.Biochem Biophys Res Commun. 2024 Dec 20;739:150952. doi: 10.1016/j.bbrc.2024.150952. Epub 2024 Nov 7. Biochem Biophys Res Commun. 2024. PMID: 39536408
-
Inhibition of Influenza Virus Polymerase by Interfering with Its Protein-Protein Interactions.ACS Infect Dis. 2021 Jun 11;7(6):1332-1350. doi: 10.1021/acsinfecdis.0c00552. Epub 2020 Oct 12. ACS Infect Dis. 2021. PMID: 33044059 Free PMC article. Review.
-
Structure-function studies of the influenza virus RNA polymerase PA subunit.Sci China C Life Sci. 2009 May;52(5):450-8. doi: 10.1007/s11427-009-0060-1. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471867 Review.
Cited by
-
Nucleocytoplasmic shuttling of influenza A virus proteins.Viruses. 2015 May 22;7(5):2668-82. doi: 10.3390/v7052668. Viruses. 2015. PMID: 26008706 Free PMC article. Review.
-
Viral and host factors required for avian H5N1 influenza A virus replication in mammalian cells.Viruses. 2013 Jun 10;5(6):1431-46. doi: 10.3390/v5061431. Viruses. 2013. PMID: 23752648 Free PMC article. Review.
-
Differential polymerase activity in avian and mammalian cells determines host range of influenza virus.J Virol. 2007 Sep;81(17):9601-4. doi: 10.1128/JVI.00666-07. Epub 2007 Jun 13. J Virol. 2007. PMID: 17567688 Free PMC article.
-
Nuclear import by karyopherin-βs: recognition and inhibition.Biochim Biophys Acta. 2011 Sep;1813(9):1593-606. doi: 10.1016/j.bbamcr.2010.10.014. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029754 Free PMC article. Review.
-
Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein.J Virol. 2009 May;83(9):4153-62. doi: 10.1128/JVI.02642-08. Epub 2009 Feb 18. J Virol. 2009. PMID: 19225007 Free PMC article.
References
-
- Bayliss, R., A. H. Corbett, and M. Stewart. 2000. The molecular mechanism of transport of macromolecules through nuclear pore complexes. Traffic 1:448-456. - PubMed
-
- Bullido, R., P. Gómez-Puertas, C. Albo, and A. Portela. 2000. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J. Gen. Virol. 81:135-142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

