Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork
- PMID: 17003056
- PMCID: PMC1635296
- DOI: 10.1093/nar/gkl677
Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork
Abstract
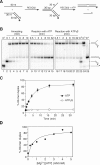
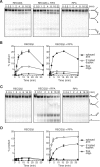
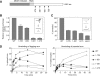
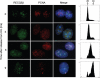
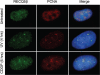
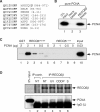
The role of the human RECQ5beta helicase in the maintenance of genomic stability remains elusive. Here we show that RECQ5beta promotes strand exchange between arms of synthetic forked DNA structures resembling a stalled replication fork in a reaction dependent on ATP hydrolysis. BLM and WRN can also promote strand exchange on these structures. However, in the presence of human replication protein A (hRPA), the action of these RecQ-type helicases is strongly biased towards unwinding of the parental duplex, an effect not seen with RECQ5beta. A domain within the non-conserved portion of RECQ5beta is identified as being important for its ability to unwind the lagging-strand arm and to promote strand exchange on hRPA-coated forked structures. We also show that RECQ5beta associates with DNA replication factories in S phase nuclei and persists at the sites of stalled replication forks after exposure of cells to UV irradiation. Moreover, RECQ5beta is found to physically interact with the polymerase processivity factor proliferating cell nuclear antigen in vitro and in vivo. Collectively, these findings suggest that RECQ5beta may promote regression of stalled replication forks to facilitate the bypass of replication-blocking lesions by template-switching. Loss of such activity could explain the elevated level of mitotic crossovers observed in RECQ5beta-deficient cells.
Figures



Similar articles
-
Escherichia coli PriA helicase: fork binding orients the helicase to unwind the lagging strand side of arrested replication forks.J Mol Biol. 2001 Oct 5;312(5):935-47. doi: 10.1006/jmbi.2001.4930. J Mol Biol. 2001. PMID: 11580240
-
RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures.Cell Cycle. 2012 Nov 15;11(22):4252-65. doi: 10.4161/cc.22581. Epub 2012 Oct 24. Cell Cycle. 2012. PMID: 23095637 Free PMC article.
-
Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide.EMBO J. 2004 Jul 21;23(14):2882-91. doi: 10.1038/sj.emboj.7600301. Epub 2004 Jul 8. EMBO J. 2004. PMID: 15241474 Free PMC article.
-
Role of the BLM helicase in replication fork management.DNA Repair (Amst). 2007 Jul 1;6(7):936-44. doi: 10.1016/j.dnarep.2007.02.007. Epub 2007 Mar 23. DNA Repair (Amst). 2007. PMID: 17363339 Review.
-
RecQ helicases: guardian angels of the DNA replication fork.Chromosoma. 2008 Jun;117(3):219-33. doi: 10.1007/s00412-007-0142-4. Epub 2008 Jan 11. Chromosoma. 2008. PMID: 18188578 Review.
Cited by
-
RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments.Genes Dev. 2007 Dec 1;21(23):3073-84. doi: 10.1101/gad.1609107. Epub 2007 Nov 14. Genes Dev. 2007. PMID: 18003859 Free PMC article.
-
Human RECQL5beta stimulates flap endonuclease 1.Nucleic Acids Res. 2010 May;38(9):2904-16. doi: 10.1093/nar/gkp1217. Epub 2010 Jan 16. Nucleic Acids Res. 2010. PMID: 20081208 Free PMC article.
-
Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding.DNA Repair (Amst). 2010 Jul 1;9(7):796-804. doi: 10.1016/j.dnarep.2010.04.003. Epub 2010 May 6. DNA Repair (Amst). 2010. PMID: 20451470 Free PMC article.
-
Cooperation of RAD51 and RAD54 in regression of a model replication fork.Nucleic Acids Res. 2011 Mar;39(6):2153-64. doi: 10.1093/nar/gkq1139. Epub 2010 Nov 21. Nucleic Acids Res. 2011. PMID: 21097884 Free PMC article.
-
Hjm/Hel308A DNA helicase from Sulfolobus tokodaii promotes replication fork regression and interacts with Hjc endonuclease in vitro.J Bacteriol. 2008 Apr;190(8):3006-17. doi: 10.1128/JB.01662-07. Epub 2008 Feb 22. J Bacteriol. 2008. PMID: 18296528 Free PMC article.
References
-
- Cox M.M. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001;35:53–82. - PubMed
-
- Cordeiro-Stone M., Makhov A.M., Zaritskaya L.S., Griffith J.D. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999;289:1207–1218. - PubMed
-
- Lopes M., Foiani M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. - PubMed
-
- Hickson I.D. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. - PubMed
-
- Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. - PubMed

