Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription
- PMID: 16990801
- PMCID: PMC1589990
- DOI: 10.1038/sj.emboj.7601332
Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription
Abstract
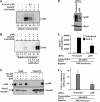
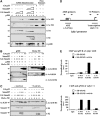
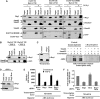
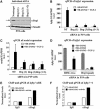
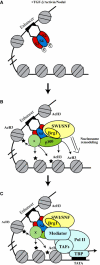
Smads are intracellular transducers for TGF-beta superfamily ligands, but little is known about the mechanism by which complexes of receptor-phosphorylated Smad2 and Smad4 regulate transcription. Using an in vitro transcription system, we have discovered that, unlike most transcription factors that are sufficient to recruit the basal transcription machinery and therefore activate transcription on both naked DNA and chromatin templates, the Smads only activate transcription from chromatin templates. We demonstrate that Smad2-mediated transcription requires the histone acetyltransferase, p300. Smad2-recruited p300 exhibits an altered substrate specificity, specifically acetylating nucleosomal histone H3 at lysines 9 and 18, and these modifications are also detected on an endogenous Smad2-dependent promoter in a ligand-induced manner. Furthermore, we show that endogenous Smad2 interacts with the SWI/SNF ATPase, Brg1, in a TGF-beta-dependent manner, and demonstrate that Brg1 is recruited to Smad2-dependent promoters and is specifically required for TGF-beta-induced expression of endogenous Smad2 target genes. Our data indicate that the Smads define a new class of transcription factors that absolutely require chromatin to assemble the basal transcription machinery and activate transcription.
Figures


Similar articles
-
Transcriptional activation by thyroid hormone receptor-beta involves chromatin remodeling, histone acetylation, and synergistic stimulation by p300 and steroid receptor coactivators.Mol Endocrinol. 2003 May;17(5):908-22. doi: 10.1210/me.2002-0308. Epub 2003 Feb 13. Mol Endocrinol. 2003. PMID: 12586842
-
Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors.J Biol Chem. 2007 May 18;282(20):14992-9. doi: 10.1074/jbc.M610316200. Epub 2007 Mar 26. J Biol Chem. 2007. PMID: 17387179 Free PMC article.
-
Acetylation of Smad2 by the co-activator p300 regulates activin and transforming growth factor beta response.J Biol Chem. 2007 Jul 20;282(29):21187-96. doi: 10.1074/jbc.M700085200. Epub 2007 May 3. J Biol Chem. 2007. PMID: 17478422
-
Chromatin remodeling during glucocorticoid receptor regulated transactivation.Biochim Biophys Acta. 2012 Jul;1819(7):716-26. doi: 10.1016/j.bbagrm.2012.02.019. Epub 2012 Mar 6. Biochim Biophys Acta. 2012. PMID: 22425674 Free PMC article. Review.
-
Intracellular signaling of the TGF-beta superfamily by Smad proteins.Ann N Y Acad Sci. 1999;886:73-82. doi: 10.1111/j.1749-6632.1999.tb09402.x. Ann N Y Acad Sci. 1999. PMID: 10667205 Review.
Cited by
-
KAP1 Is a Chromatin Reader that Couples Steps of RNA Polymerase II Transcription to Sustain Oncogenic Programs.Mol Cell. 2020 Jun 18;78(6):1133-1151.e14. doi: 10.1016/j.molcel.2020.04.024. Epub 2020 May 12. Mol Cell. 2020. PMID: 32402252 Free PMC article.
-
Master transcription factors determine cell-type-specific responses to TGF-β signaling.Cell. 2011 Oct 28;147(3):565-76. doi: 10.1016/j.cell.2011.08.050. Cell. 2011. PMID: 22036565 Free PMC article.
-
Differential kinase activity of ACVR1 G328V and R206H mutations with implications to possible TβRI cross-talk in diffuse intrinsic pontine glioma.Sci Rep. 2020 Apr 9;10(1):6140. doi: 10.1038/s41598-020-63061-0. Sci Rep. 2020. PMID: 32273545 Free PMC article.
-
The BMP Pathway in Blood Vessel and Lymphatic Vessel Biology.Int J Mol Sci. 2021 Jun 14;22(12):6364. doi: 10.3390/ijms22126364. Int J Mol Sci. 2021. PMID: 34198654 Free PMC article. Review.
-
A single-cell sequence analysis of mouse subcutaneous white adipose tissue reveals dynamic changes during weaning.Commun Biol. 2024 Jun 29;7(1):787. doi: 10.1038/s42003-024-06448-3. Commun Biol. 2024. PMID: 38951550 Free PMC article.
References
-
- Alberts AS, Geneste O, Treisman R (1998) Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell 92: 475–487 - PubMed
-
- Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K (2004) Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol Cell 15: 813–823 - PubMed
-
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW (2005) The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci 30: 250–255 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

