Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling
- PMID: 16987958
- PMCID: PMC1679666
- DOI: 10.1091/mbc.e06-06-0513
Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling
Abstract
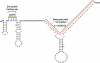
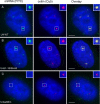
Spliceosomal small nuclear ribonucleoprotein particles (snRNPs) undergo specific assembly steps in Cajal bodies (CBs), nonmembrane-bound compartments within cell nuclei. An example is the U4/U6 di-snRNP, assembled from U4 and U6 monomers. These snRNPs can also assemble in the nucleoplasm when cells lack CBs. Here, we address the hypothesis that snRNP concentration in CBs facilitates assembly, by comparing the predicted rates of U4 and U6 snRNP association in nuclei with and without CBs. This was accomplished by a random walk-and-capture simulation applied to a three-dimensional model of the HeLa cell nucleus, derived from measurements of living cells. Results of the simulations indicated that snRNP capture is optimal when nuclei contain three to four CBs. Interestingly, this is the observed number of CBs in most cells. Microinjection experiments showed that U4 snRNA targeting to CBs was U6 snRNP independent and that snRNA concentration in CBs is approximately 20-fold higher than in nucleoplasm. Finally, combination of the simulation with calculated association rates predicted that the presence of CBs enhances U4 and U6 snRNP association by up to 11-fold, largely owing to this concentration difference. This provides a chemical foundation for the proposal that these and other cellular compartments promote molecular interactions, by increasing the local concentration of individual components.
Figures
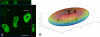


Similar articles
-
Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies.J Cell Biol. 2003 Feb 17;160(4):505-16. doi: 10.1083/jcb.200210087. Epub 2003 Feb 10. J Cell Biol. 2003. PMID: 12578909 Free PMC article.
-
Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer.J Cell Biol. 2004 Sep 27;166(7):1015-25. doi: 10.1083/jcb.200405160. J Cell Biol. 2004. PMID: 15452143 Free PMC article.
-
RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies.EMBO J. 2004 Aug 4;23(15):3000-9. doi: 10.1038/sj.emboj.7600296. Epub 2004 Jul 15. EMBO J. 2004. PMID: 15257298 Free PMC article.
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
-
The assembly of a spliceosomal small nuclear ribonucleoprotein particle.Nucleic Acids Res. 2008 Nov;36(20):6482-93. doi: 10.1093/nar/gkn658. Epub 2008 Oct 14. Nucleic Acids Res. 2008. PMID: 18854356 Free PMC article. Review.
Cited by
-
Small-molecule flunarizine increases SMN protein in nuclear Cajal bodies and motor function in a mouse model of spinal muscular atrophy.Sci Rep. 2018 Feb 1;8(1):2075. doi: 10.1038/s41598-018-20219-1. Sci Rep. 2018. PMID: 29391529 Free PMC article.
-
The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation.Nucleic Acids Res. 2018 Apr 20;46(7):3774-3790. doi: 10.1093/nar/gky070. Nucleic Acids Res. 2018. PMID: 29415178 Free PMC article.
-
Do Cellular Condensates Accelerate Biochemical Reactions? Lessons from Microdroplet Chemistry.Biophys J. 2018 Jul 3;115(1):3-8. doi: 10.1016/j.bpj.2018.05.023. Biophys J. 2018. PMID: 29972809 Free PMC article. Review.
-
TDP-43 and NEAT long non-coding RNA: Roles in neurodegenerative disease.Front Cell Neurosci. 2022 Oct 26;16:954912. doi: 10.3389/fncel.2022.954912. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36385948 Free PMC article. Review.
-
P-bodies and stress granules: possible roles in the control of translation and mRNA degradation.Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a012286. doi: 10.1101/cshperspect.a012286. Cold Spring Harb Perspect Biol. 2012. PMID: 22763747 Free PMC article. Review.
References
-
- Berg H. Random Walks in Biology. Princeton, NJ: Princeton University Press; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

