A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer
- PMID: 16980581
- PMCID: PMC1578675
- DOI: 10.1101/gad.1446006
A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer
Abstract
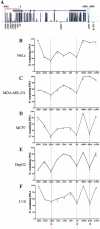
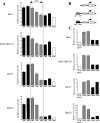
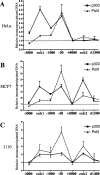
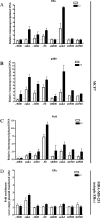
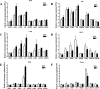
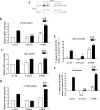
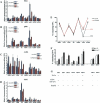
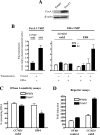
Estrogen stimulates the proliferation of the most common type of human breast cancer that expresses estrogen receptor alpha (ERalpha) through the activation of the cyclin D1 (CCND1) oncogene. However, our knowledge of ERalpha transcriptional mechanisms remains limited. Hence, it is still elusive why ERalpha ectopically expressed in ER-negative breast cancer cells (BCC) is functional on ectopic reporter constructs but lacks activity on many endogenous target genes, including CCND1. Here, we show that estradiol (E2) stimulation of CCND1 expression in BCC depends on a novel cell-type-specific enhancer downstream from the CCND1 coding region, which is the primary ERalpha recruitment site in estrogen-responsive cells. The pioneer factor FoxA1 is specifically required for the active chromatin state of this enhancer and as such is crucial for both CCND1 expression and subsequent cell cycle progression. Interestingly, even in BCC, CCND1 levels and proliferation are tightly controlled by E2 through the establishment of a negative feedforward loop involving the induction of NFIC, a putative tumor suppressor capable of directly repressing CCND1 transcription. Taken together, our results reveal an estrogen-regulated combinatorial network including cell-specific cis- and trans-regulators of CCND1 expression where ERalpha collaborates with other transcription factors associated with the ER-positive breast cancer phenotype, including FoxA1 and NFIC.
Figures
Similar articles
-
Estrogen receptor alpha mediates progestin-induced mammary tumor growth by interacting with progesterone receptors at the cyclin D1/MYC promoters.Cancer Res. 2012 May 1;72(9):2416-27. doi: 10.1158/0008-5472.CAN-11-3290. Epub 2012 Mar 6. Cancer Res. 2012. PMID: 22396492
-
From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response.Proc Natl Acad Sci U S A. 2005 Aug 16;102(33):11651-6. doi: 10.1073/pnas.0505575102. Epub 2005 Aug 8. Proc Natl Acad Sci U S A. 2005. PMID: 16087863 Free PMC article.
-
Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity.Cancer Res. 2005 Aug 1;65(15):6557-67. doi: 10.1158/0008-5472.CAN-05-0486. Cancer Res. 2005. PMID: 16061635
-
Role of RUNX2 in Breast Carcinogenesis.Int J Mol Sci. 2015 Sep 2;16(9):20969-93. doi: 10.3390/ijms160920969. Int J Mol Sci. 2015. PMID: 26404249 Free PMC article. Review.
-
FOXA1 as a therapeutic target for breast cancer.Expert Opin Ther Targets. 2007 Apr;11(4):507-14. doi: 10.1517/14728222.11.4.507. Expert Opin Ther Targets. 2007. PMID: 17373880 Review.
Cited by
-
Cyclin D1 determines estrogen signaling in the mammary gland in vivo.Mol Endocrinol. 2013 Sep;27(9):1415-28. doi: 10.1210/me.2013-1065. Epub 2013 Jul 17. Mol Endocrinol. 2013. PMID: 23864650 Free PMC article.
-
In-silico identification and functional validation of allele-dependent AR enhancers.Oncotarget. 2015 Mar 10;6(7):4816-28. doi: 10.18632/oncotarget.3019. Oncotarget. 2015. PMID: 25693204 Free PMC article.
-
Inferring regulatory element landscapes and transcription factor networks from cancer methylomes.Genome Biol. 2015 May 21;16(1):105. doi: 10.1186/s13059-015-0668-3. Genome Biol. 2015. PMID: 25994056 Free PMC article.
-
PES1 promotes breast cancer by differentially regulating ERα and ERβ.J Clin Invest. 2012 Aug;122(8):2857-70. doi: 10.1172/JCI62676. Epub 2012 Jul 23. J Clin Invest. 2012. PMID: 22820289 Free PMC article.
-
The Central Contributions of Breast Cancer Stem Cells in Developing Resistance to Endocrine Therapy in Estrogen Receptor (ER)-Positive Breast Cancer.Cancers (Basel). 2019 Jul 22;11(7):1028. doi: 10.3390/cancers11071028. Cancers (Basel). 2019. PMID: 31336602 Free PMC article. Review.
References
-
- Altucci, L., Addeo, R., Cicatiello, L., Dauvois, S., Parker, M.G., Truss, M., Beato, M., Sica, V., Bresciani, F., Weisz, A. 17β-Estradiol induces cyclin D1 gene transcription, p36D1–p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G(1)-arrested human breast cancer cells. Oncogene. 1996;12:2315–2324. - PubMed
-
- Arnold, A., Papanikolaou, A. Cyclin D1 in breast cancer pathogenesis. J. Clin. Oncol. 2005;23:4215–4224. - PubMed
-
- Arnosti, D.N., Kulkarni, M.M. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 2005;94:890–898. - PubMed
-
- Carroll, J.S., Liu, X.S., Brodsky, A.S., Li, W., Meyer, C.A., Szary, A.J., Eeckhoute, J., Shao, W., Hestermann, E.V., Geistlinger, T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Castro-Rivera, E., Samudio, I., Safe, S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 2001;276:30853–30861. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
