Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-kappaB binding sites in the enhancer
- PMID: 16973595
- PMCID: PMC1617225
- DOI: 10.1128/JVI.00640-06
Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-kappaB binding sites in the enhancer
Abstract
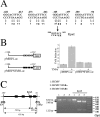
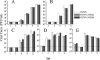
The role of NF-kappaB in regulating human cytomegalovirus (HCMV) replication and gene transcription remains controversial. Multiple, functional NF-kappaB response elements exist in the major immediate-early promoter (MIEP) enhancer of HCMV, suggesting a possible requirement for this transcription factor in lytic viral replication. Here we demonstrate by generating and analyzing HCMVs with alterations in the MIEP-enhancer that, although this region is essential for HCMV growth, none of the four NF-kappaB response elements contained within the enhancer are required for MIE gene expression or HCMV replication in multiple cell types. These data reveal the robustness of the regulatory network controlling the MIEP enhancer.
Figures

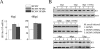
Similar articles
-
The Elk-1 and serum response factor binding sites in the major immediate-early promoter of human cytomegalovirus are required for efficient viral replication in quiescent cells and compensate for inactivation of the NF-kappaB sites in proliferating cells.J Virol. 2010 May;84(9):4481-93. doi: 10.1128/JVI.02141-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147408 Free PMC article.
-
Activation of the virus-induced IKK/NF-kappaB signalling axis is critical for the replication of human cytomegalovirus in quiescent cells.Cell Microbiol. 2007 Aug;9(8):2040-54. doi: 10.1111/j.1462-5822.2007.00936.x. Epub 2007 Apr 5. Cell Microbiol. 2007. PMID: 17419715
-
Two gamma interferon-activated site-like elements in the human cytomegalovirus major immediate-early promoter/enhancer are important for viral replication.J Virol. 2005 Apr;79(8):5035-46. doi: 10.1128/JVI.79.8.5035-5046.2005. J Virol. 2005. PMID: 15795289 Free PMC article.
-
Immediate-early transcription regulation of human cytomegalovirus.Curr Top Microbiol Immunol. 1990;154:3-19. doi: 10.1007/978-3-642-74980-3_1. Curr Top Microbiol Immunol. 1990. PMID: 2161324 Review. No abstract available.
-
Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency.Med Microbiol Immunol. 2008 Jun;197(2):223-31. doi: 10.1007/s00430-007-0069-7. Epub 2007 Dec 19. Med Microbiol Immunol. 2008. PMID: 18097687 Review.
Cited by
-
Phorbol ester-induced human cytomegalovirus major immediate-early (MIE) enhancer activation through PKC-delta, CREB, and NF-kappaB desilences MIE gene expression in quiescently infected human pluripotent NTera2 cells.J Virol. 2010 Sep;84(17):8495-508. doi: 10.1128/JVI.00416-10. Epub 2010 May 26. J Virol. 2010. PMID: 20504934 Free PMC article.
-
New cell-signaling pathways for controlling cytomegalovirus replication.Am J Transplant. 2014 Jun;14(6):1249-58. doi: 10.1111/ajt.12725. Epub 2014 May 19. Am J Transplant. 2014. PMID: 24839861 Free PMC article. Review.
-
cGAS-STING-TBK1 Signaling Promotes Valproic Acid-Responsive Human Cytomegalovirus Immediate-Early Transcription during Infection of Incompletely Differentiated Myeloid Cells.Viruses. 2024 May 30;16(6):877. doi: 10.3390/v16060877. Viruses. 2024. PMID: 38932169 Free PMC article.
-
Proinflammatory cytokine-induced tight junction remodeling through dynamic self-assembly of claudins.Mol Biol Cell. 2014 Sep 15;25(18):2710-9. doi: 10.1091/mbc.E14-02-0773. Epub 2014 Jul 16. Mol Biol Cell. 2014. PMID: 25031428 Free PMC article.
-
NFκB and Cyclic AMP Response Element Sites Mediate the Valproic Acid and UL138 Responsiveness of the Human Cytomegalovirus Major Immediate Early Enhancer and Promoter.J Virol. 2023 Mar 30;97(3):e0002923. doi: 10.1128/jvi.00029-23. Epub 2023 Mar 1. J Virol. 2023. PMID: 36856444 Free PMC article.
References
-
- Benedict, C. A., T. A. Banks, L. Senderowicz, M. Ko, W. J. Britt, A. Angulo, P. Ghazal, and C. F. Ware. 2001. Lymphotoxins and cytomegalovirus cooperatively induce interferon-beta, establishing host-virus detente. Immunity 15:617-626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

