Identification of a B-cell antigenic epitope at the N-terminus of SARS-CoV M protein and characterization of monoclonal antibody against the protein
- PMID: 16972028
- PMCID: PMC7088559
- DOI: 10.1007/s11262-005-0050-8
Identification of a B-cell antigenic epitope at the N-terminus of SARS-CoV M protein and characterization of monoclonal antibody against the protein
Abstract
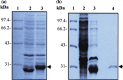
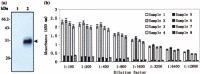
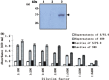
To identify the potential B-cell antigenic epitopes within the N-terminus of SARS-CoV (SARS-associated coronavirus, SARS-CoV) M protein and characterize monoclonal antibody (MAb) against the protein as well as its recognizing region, we expressed and purified a portion of SARS-CoV M protein (amino acid 1-43) in Escherichia coli (E. coli). By using Western blot and enzyme-linked immunosorbent assay (ELISA), we showed that the purified recombinant M protein could be recognized by four SARS-CoV-positive human sera even when those sera were 12,800-fold diluted. Furthermore, we characterized one representative IgG2 MAb, 3H9, which exhibited a strong immunoreaction to both recombinant M protein and native viral protein of SARS-CoV. We found a B-cell antigenic epitope located between amino acid 1-15 and defined the MAb recognizing region within amino acid 16-28 of M. These findings not only suggest that both recombinant M protein and its specific MAbs may be used as the diagnostic reagents for SARS, but also provide a potential target site for the design of an epitope-based vaccine against SARS.
Figures


Similar articles
-
A chimeric multi-epitope DNA vaccine elicited specific antibody response against severe acute respiratory syndrome-associated coronavirus which attenuated the virulence of SARS-CoV in vitro.Immunol Lett. 2008 Aug 15;119(1-2):71-7. doi: 10.1016/j.imlet.2008.04.005. Epub 2008 May 19. Immunol Lett. 2008. PMID: 18533276 Free PMC article.
-
Monoclonal antibodies to SARS-associated coronavirus (SARS-CoV): identification of neutralizing and antibodies reactive to S, N, M and E viral proteins.J Virol Methods. 2005 Sep;128(1-2):21-8. doi: 10.1016/j.jviromet.2005.03.021. J Virol Methods. 2005. PMID: 15885812 Free PMC article.
-
Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA.J Med Virol. 2004 Jul;73(3):338-46. doi: 10.1002/jmv.20096. J Med Virol. 2004. PMID: 15170626 Free PMC article.
-
Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology.MAbs. 2010 Jan-Feb;2(1):53-66. doi: 10.4161/mabs.2.1.10788. Epub 2010 Jan 27. MAbs. 2010. PMID: 20168090 Free PMC article.
-
Two-way antigenic cross-reactivity between severe acute respiratory syndrome coronavirus (SARS-CoV) and group 1 animal CoVs is mediated through an antigenic site in the N-terminal region of the SARS-CoV nucleoprotein.J Virol. 2007 Dec;81(24):13365-77. doi: 10.1128/JVI.01169-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913799 Free PMC article.
Cited by
-
Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus.Virol J. 2012 Oct 1;9:225. doi: 10.1186/1743-422X-9-225. Virol J. 2012. PMID: 23025700 Free PMC article.
-
Predicted 3D model of the M protein of Porcine Epidemic Diarrhea Virus and analysis of its immunogenic potential.PLoS One. 2022 Feb 9;17(2):e0263582. doi: 10.1371/journal.pone.0263582. eCollection 2022. PLoS One. 2022. PMID: 35139120 Free PMC article.
-
A Candidate Antigen of the Recombinant Membrane Protein Derived from the Porcine Deltacoronavirus Synthetic Gene to Detect Seropositive Pigs.Viruses. 2023 Apr 25;15(5):1049. doi: 10.3390/v15051049. Viruses. 2023. PMID: 37243136 Free PMC article.
-
Development and Application of an Indirect Enzyme-Linked Immunosorbent Assay Based on a Recombinant Matrix Protein for the Serological Study of Porcine Deltacoronavirus in Mexican Pigs.Vet Med Sci. 2024 Nov;10(6):e70108. doi: 10.1002/vms3.70108. Vet Med Sci. 2024. PMID: 39494986 Free PMC article.
-
Recent advances in immunoassay technologies for the detection of human coronavirus infections.Front Cell Infect Microbiol. 2023 Jan 4;12:1040248. doi: 10.3389/fcimb.2022.1040248. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36683684 Free PMC article. Review.
References
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

