Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells
- PMID: 16971455
- PMCID: PMC1635286
- DOI: 10.1093/nar/gkl646
Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells
Abstract
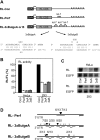
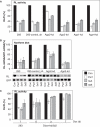
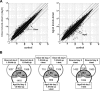
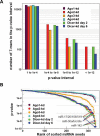
RNA interference and the microRNA (miRNA) pathway can induce sequence-specific mRNA degradation and/or translational repression. The human genome encodes hundreds of miRNAs that can post-transcriptionally repress thousands of genes. Using reporter constructs, we observed that degradation of mRNAs bearing sites imperfectly complementary to the endogenous let-7 miRNA is considerably stronger in human HEK293 than HeLa cells. The degradation did not result from the Ago2-mediated endonucleolytic cleavage but it was Dicer- and Ago2-dependent. We used this feature of HEK293 to address the size of a pool of transcripts regulated by RNA silencing in a single cell type. We generated HEK293 cell lines depleted of Dicer or individual Ago proteins. The cell lines were used for microarray analyses to obtain a comprehensive picture of RNA silencing. The 3'-untranslated region sequences of a few hundred transcripts that were commonly up-regulated upon Ago2 and Dicer knock-downs showed a significant enrichment of putative miRNA-binding sites. The up-regulation upon Ago2 and Dicer knock-downs was moderate and we found no evidence, at the mRNA level, for activation of silenced genes. Taken together, our data suggest that, independent of the effect on translation, miRNAs affect levels of a few hundred mRNAs in HEK293 cells.
Figures


Similar articles
-
Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54.PLoS Biol. 2006 Jul;4(7):e210. doi: 10.1371/journal.pbio.0040210. PLoS Biol. 2006. PMID: 16756390 Free PMC article.
-
TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing.Nature. 2005 Aug 4;436(7051):740-4. doi: 10.1038/nature03868. Epub 2005 Jun 22. Nature. 2005. PMID: 15973356 Free PMC article.
-
Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster.Mol Cell Biol. 2006 Apr;26(8):2965-75. doi: 10.1128/MCB.26.8.2965-2975.2006. Mol Cell Biol. 2006. PMID: 16581772 Free PMC article.
-
A brief history of RNAi: the silence of the genes.FASEB J. 2006 Jul;20(9):1293-9. doi: 10.1096/fj.06-6014rev. FASEB J. 2006. PMID: 16816104 Review.
-
miRNPs: versatile regulators of gene expression in vertebrate cells.Biochem Soc Trans. 2009 Oct;37(Pt 5):931-5. doi: 10.1042/BST0370931. Biochem Soc Trans. 2009. PMID: 19754429 Review.
Cited by
-
Context-dependent regulation of Dicer activity and small RNA production: Implications to oocyte-to-embryo transition.Worm. 2015 Sep 11;4(4):e1086062. doi: 10.1080/21624054.2015.1086062. eCollection 2015 Oct-Dec. Worm. 2015. PMID: 27123367 Free PMC article.
-
DMRT1 repression using a novel approach to genetic manipulation induces testicular dysgenesis in human fetal gonads.Hum Reprod. 2018 Nov 1;33(11):2107-2121. doi: 10.1093/humrep/dey289. Hum Reprod. 2018. PMID: 30272154 Free PMC article.
-
MicroRNA activity is suppressed in mouse oocytes.Curr Biol. 2010 Feb 9;20(3):265-70. doi: 10.1016/j.cub.2009.12.042. Epub 2010 Jan 28. Curr Biol. 2010. PMID: 20116252 Free PMC article.
-
Determinants of targeting by endogenous and exogenous microRNAs and siRNAs.RNA. 2007 Nov;13(11):1894-910. doi: 10.1261/rna.768207. Epub 2007 Sep 13. RNA. 2007. PMID: 17872505 Free PMC article.
-
Physiologically relevant miRNAs in mammalian oocytes are rare and highly abundant.EMBO Rep. 2022 Feb 3;23(2):e53514. doi: 10.15252/embr.202153514. Epub 2021 Dec 6. EMBO Rep. 2022. PMID: 34866300 Free PMC article.
References
-
- Sontheimer E.J., Carthew R.W. Silence from within: endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. - PubMed
-
- Zamore P.D., Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. - PubMed
-
- Marques J.T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M.A., Williams B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

