Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products
- PMID: 16971431
- PMCID: PMC1642625
- DOI: 10.1128/JVI.00940-06
Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products
Abstract
Acanthamoeba polyphaga mimivirus is the largest known virus in both particle size and genome complexity. Its 1.2-Mb genome encodes 911 proteins, among which only 298 have predicted functions. The composition of purified isolated virions was analyzed by using a combined electrophoresis/mass spectrometry approach allowing the identification of 114 proteins. Besides the expected major structural components, the viral particle packages 12 proteins unambiguously associated with transcriptional machinery, 3 proteins associated with DNA repair, and 2 topoisomerases. Other main functional categories represented in the virion include oxidative pathways and protein modification. More than half of the identified virion-associated proteins correspond to anonymous genes of unknown function, including 45 "ORFans." As demonstrated by both Western blotting and immunogold staining, some of these "ORFans," which lack any convincing similarity in the sequence databases, are endowed with antigenic properties. Thus, anonymous and unique genes constituting the majority of the mimivirus gene complement encode bona fide proteins that are likely to participate in well-integrated processes.
Figures
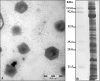


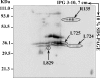
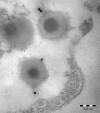
Similar articles
-
The 1.2-megabase genome sequence of Mimivirus.Science. 2004 Nov 19;306(5700):1344-50. doi: 10.1126/science.1101485. Epub 2004 Oct 14. Science. 2004. PMID: 15486256
-
Structural and functional insights into Mimivirus ORFans.BMC Genomics. 2007 May 9;8:115. doi: 10.1186/1471-2164-8-115. BMC Genomics. 2007. PMID: 17490476 Free PMC article.
-
The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription and DNA repair.Virology. 2014 Oct;466-467:82-94. doi: 10.1016/j.virol.2014.05.029. Epub 2014 Jun 25. Virology. 2014. PMID: 24973308
-
Mimivirus and the emerging concept of "giant" virus.Virus Res. 2006 Apr;117(1):133-44. doi: 10.1016/j.virusres.2006.01.008. Epub 2006 Feb 15. Virus Res. 2006. PMID: 16469402 Review.
-
Gene repertoire of amoeba-associated giant viruses.Intervirology. 2010;53(5):330-43. doi: 10.1159/000312918. Epub 2010 Jun 15. Intervirology. 2010. PMID: 20551685 Review.
Cited by
-
Exploring ORFan domains in giant viruses: structure of mimivirus sulfhydryl oxidase R596.PLoS One. 2012;7(11):e50649. doi: 10.1371/journal.pone.0050649. Epub 2012 Nov 28. PLoS One. 2012. PMID: 23209798 Free PMC article.
-
Vaccinia-like cytoplasmic replication of the giant Mimivirus.Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):5978-82. doi: 10.1073/pnas.0912737107. Epub 2010 Mar 15. Proc Natl Acad Sci U S A. 2010. PMID: 20231474 Free PMC article.
-
Host genome integration and giant virus-induced reactivation of the virophage mavirus.Nature. 2016 Dec 7;540(7632):288-291. doi: 10.1038/nature20593. Nature. 2016. PMID: 27929021
-
Lamarckian evolution of the giant Mimivirus in allopatric laboratory culture on amoebae.Front Cell Infect Microbiol. 2012 Jul 5;2:91. doi: 10.3389/fcimb.2012.00091. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919682 Free PMC article.
-
Widespread Distribution and Evolution of Poxviral Entry-Fusion Complex Proteins in Giant Viruses.Microbiol Spectr. 2023 Mar 13;11(2):e0494422. doi: 10.1128/spectrum.04944-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36912656 Free PMC article.
References
-
- Amiri, H., W. Davids, and S. G. Andersson. 2003. Birth and death of orphan genes in Rickettsia. Mol. Biol. Evol. 20:1575-1587. - PubMed
-
- Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 16:673-685. - PubMed
-
- Bustamante, C., Z. Bryant, and S. B. Smith. 2003. Ten years of tension: single-molecule DNA mechanics. Nature 421:423-427. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

