The C-terminal hydrophobic domain of hepatitis C virus RNA polymerase NS5B can be replaced with a heterologous domain of poliovirus protein 3A
- PMID: 16971430
- PMCID: PMC1642141
- DOI: 10.1128/JVI.02072-05
The C-terminal hydrophobic domain of hepatitis C virus RNA polymerase NS5B can be replaced with a heterologous domain of poliovirus protein 3A
Abstract
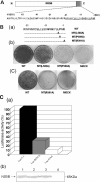
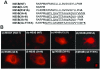
Replication of the plus-stranded RNA genome of hepatitis C virus (HCV) occurs in a membrane-bound replication complex consisting of viral and cellular proteins and viral RNA. NS5B, the RNA polymerase of HCV, is anchored to the membranes via a C-terminal 20-amino-acid-long hydrophobic domain, which is flanked on each side by a highly conserved positively charged arginine. Using a genotype 1b subgenomic replicon (V. Lohmann, F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartensclager, Science 285:110-113, 1999), we determined the effect of mutations of some highly conserved residues in this domain. The replacement of arginine 570 with alanine completely abolished the colony-forming ability by the replicon, while a R591A change was found to be highly detrimental to replication, viability, and membrane binding by the mutant NS5B protein. Mutations of two other highly conserved amino acids (L588A and P589A) reduced but did not eliminate colony formation. It was of interest, if specific amino acid residues play a role in membrane anchoring of NS5B and replication, to determine whether a complete exchange of the NS5B hydrophobic domain with a domain totally unrelated to NS5B would ablate replication. We selected the 22-amino-acid-long hydrophobic domain of poliovirus polypeptide 3A that is known to adopt a transmembrane configuration, thereby anchoring 3A to membranes. Surprisingly, either partial or full replacement of the NS5B hydrophobic domain with the anchor sequences of poliovirus polypeptide 3A resulted in the replication of replicons whose colony-forming abilities were reduced compared to that of the wild-type replicon. Upon continued passage of the replicon in Huh-7 cells in the presence of neomycin, the replication efficiency of the replicon increased. However, the sequence of the poliovirus polypeptide 3A hydrophobic domain, in the context of the subgenomic HCV replicon, was stably maintained throughout 40 passages. Our results suggest that anchoring NS5B to membranes is necessary but that the amino acid sequence of the anchor per se does not require HCV origin. This suggests that specific interactions between the NS5B hydrophobic domain and other membrane-bound factors may not play a decisive role in HCV replication.
Figures


Similar articles
-
Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture.Virology. 2002 Jun 5;297(2):298-306. doi: 10.1006/viro.2002.1461. Virology. 2002. PMID: 12083828
-
The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo.J Virol. 2004 Apr;78(7):3797-802. doi: 10.1128/jvi.78.7.3797-3802.2004. J Virol. 2004. PMID: 15016899 Free PMC article.
-
Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication.J Virol. 2004 May;78(10):5288-98. doi: 10.1128/jvi.78.10.5288-5298.2004. J Virol. 2004. PMID: 15113910 Free PMC article.
-
[Interferon resistance and ISDR (interferon sensitivity determining region)].Nihon Rinsho. 2006 Jul;64(7):1249-53. Nihon Rinsho. 2006. PMID: 16838640 Review. Japanese.
-
Nonstructural protein 5B of hepatitis C virus.Mol Cells. 2006 Jun 30;21(3):330-6. Mol Cells. 2006. PMID: 16819294 Review.
Cited by
-
Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome.J Virol. 2013 May;87(10):5678-96. doi: 10.1128/JVI.00840-12. Epub 2013 Mar 13. J Virol. 2013. PMID: 23487449 Free PMC article.
-
Hepatitis C virus RNA replication requires a conserved structural motif within the transmembrane domain of the NS5B RNA-dependent RNA polymerase.J Virol. 2010 Nov;84(21):11580-4. doi: 10.1128/JVI.01519-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739529 Free PMC article.
-
Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication.Biochemistry. 2007 May 1;46(17):5185-99. doi: 10.1021/bi6024758. Epub 2007 Apr 7. Biochemistry. 2007. PMID: 17417822 Free PMC article.
References
-
- Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. - PubMed
-
- Aizaki, H., K. J. Lee, V. M. H. Sung, H. Ishiko, and M. M. C. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450-461. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic α-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

