The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles
- PMID: 16963142
- PMCID: PMC2676329
- DOI: 10.1016/j.virusres.2006.08.003
The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles
Abstract
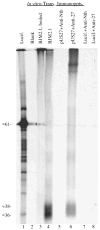
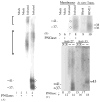
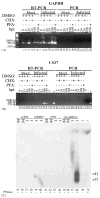
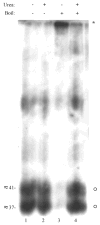
Human cytomegalovirus (HCMV), a member of the beta-herpesvirus family, encodes four homologues of cellular G protein-coupled receptors (GPCRs). One of these, the protein product of HCMV open reading frame (ORF) UL33, has been identified in HCMV-infected cells and virus particles and shown to be heat-aggregatable and N-glycosylated. Another, the product of ORF US28, has been functionally characterized as a beta-chemokine receptor. Here we report the use of RT-PCR, coupled in vitro transcription-translation, immunoprecipitation, and Western immunoassays to (i) show that RNA from the open reading frame US27 appears predominantly during the late phase of replication; (ii) identify the protein encoded by HCMV US27 in infected cells and enveloped virus particles; (iii) demonstrate that the US27-encoded protein is heterogeneously N-glycosylated and resolves as two species following treatment with peptide N-glycosidase F; and (iv) show that both the recombinant and deglycoylated infected cell US27 protein aggregate when heated in the presence of SDS prior to electrophoresis in polyacrylamide gels, a property which is abrogated with the addition of urea to sample buffer.
Figures

Similar articles
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles.Virology. 1996 Nov 1;225(1):111-25. doi: 10.1006/viro.1996.0579. Virology. 1996. PMID: 8918538 Free PMC article.
-
Human Cytomegalovirus UL111A and US27 Gene Products Enhance the CXCL12/CXCR4 Signaling Axis via Distinct Mechanisms.J Virol. 2018 Feb 12;92(5):e01981-17. doi: 10.1128/JVI.01981-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237840 Free PMC article.
-
The human cytomegalovirus chemokine receptor homolog encoded by US27.Virus Genes. 2017 Aug;53(4):516-521. doi: 10.1007/s11262-017-1462-y. Epub 2017 Apr 26. Virus Genes. 2017. PMID: 28447191 Review.
-
Emerging roles of cytomegalovirus-encoded G protein-coupled receptors during lytic and latent infection.Med Microbiol Immunol. 2019 Aug;208(3-4):447-456. doi: 10.1007/s00430-019-00595-9. Epub 2019 Mar 21. Med Microbiol Immunol. 2019. PMID: 30900091 Review.
Cited by
-
Identification of a novel signaling complex containing host chemokine receptor CXCR4, Interleukin-10 receptor, and human cytomegalovirus US27.Virology. 2020 Sep;548:49-58. doi: 10.1016/j.virol.2020.06.006. Epub 2020 Jun 17. Virology. 2020. PMID: 32838946 Free PMC article.
-
Mechanisms of Regulation of the Chemokine-Receptor Network.Int J Mol Sci. 2017 Feb 7;18(2):342. doi: 10.3390/ijms18020342. Int J Mol Sci. 2017. PMID: 28178200 Free PMC article. Review.
-
Intracellular trafficking of the human cytomegalovirus-encoded 7-trans-membrane protein homologs pUS27 and pUL78 during viral infection: a comparative analysis.Viruses. 2014 Feb 10;6(2):661-82. doi: 10.3390/v6020661. Viruses. 2014. PMID: 24517969 Free PMC article.
-
Methods for Studying the Function of Cytomegalovirus GPCRs.Methods Mol Biol. 2021;2244:159-197. doi: 10.1007/978-1-0716-1111-1_9. Methods Mol Biol. 2021. PMID: 33555587
-
Desensitization of herpesvirus-encoded G protein-coupled receptors.Life Sci. 2008 Jan 16;82(3-4):125-34. doi: 10.1016/j.lfs.2007.10.024. Epub 2007 Nov 13. Life Sci. 2008. PMID: 18054964 Free PMC article. Review.
References
-
- AbuBakar S, Boldogh I, Albrecht T. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholipase A2 and protein kinase C. Biochem Biophys Res Commun. 1990a;166(2):953–959. - PubMed
-
- AbuBakar S, Boldogh I, Albrecht T. Human cytomegalovirus: stimulation of [3H] release from [3H]-arachidonic acid prelabelled cells. Arch Virol. 1990b;113:255–266. - PubMed
-
- Albrecht T, Fons MP, Boldogh I, AbuBakar S, Deng CZ, Millinoff D. Metabolic and cellular effects of human cytomegalovirus infection. Transplant Proc. 1991;23(3):48S–55S. - PubMed
-
- Attwood TK, Findlay JBC. Design of a discriminating fingerprint for G-protein-coupled receptors. Protein Eng. 1994a;6(2):167–176. - PubMed
-
- Attwood TK, Findlay JBC. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994b;7(2):195–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

