Statistical tests for detecting positive selection by utilizing high-frequency variants
- PMID: 16951063
- PMCID: PMC1667063
- DOI: 10.1534/genetics.106.061432
Statistical tests for detecting positive selection by utilizing high-frequency variants
Abstract
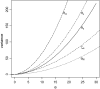
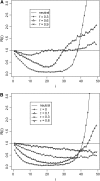
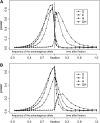
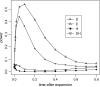
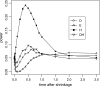
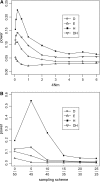
By comparing the low-, intermediate-, and high-frequency parts of the frequency spectrum, we gain information on the evolutionary forces that influence the pattern of polymorphism in population samples. We emphasize the high-frequency variants on which positive selection and negative (background) selection exhibit different effects. We propose a new estimator of theta (the product of effective population size and neutral mutation rate), thetaL, which is sensitive to the changes in high-frequency variants. The new thetaL allows us to revise Fay and Wu's H-test by normalization. To complement the existing statistics (the H-test and Tajima's D-test), we propose a new test, E, which relies on the difference between thetaL and Watterson's thetaW. We show that this test is most powerful in detecting the recovery phase after the loss of genetic diversity, which includes the postselective sweep phase. The sensitivities of these tests to (or robustness against) background selection and demographic changes are also considered. Overall, D and H in combination can be most effective in detecting positive selection while being insensitive to other perturbations. We thus propose a joint test, referred to as the DH test. Simulations indicate that DH is indeed sensitive primarily to directional selection and no other driving forces.
Figures
Similar articles
-
Compound tests for the detection of hitchhiking under positive selection.Mol Biol Evol. 2007 Aug;24(8):1898-908. doi: 10.1093/molbev/msm119. Epub 2007 Jun 8. Mol Biol Evol. 2007. PMID: 17557886
-
A new test for detecting recent positive selection that is free from the confounding impacts of demography.Mol Biol Evol. 2011 Jan;28(1):365-75. doi: 10.1093/molbev/msq211. Epub 2010 Aug 13. Mol Biol Evol. 2011. PMID: 20709734
-
Neutrality tests for sequences with missing data.Genetics. 2012 Aug;191(4):1397-401. doi: 10.1534/genetics.112.139949. Epub 2012 Jun 1. Genetics. 2012. PMID: 22661328 Free PMC article.
-
Detecting directional selection in the presence of recent admixture in African-Americans.Genetics. 2011 Mar;187(3):823-35. doi: 10.1534/genetics.110.122739. Epub 2010 Dec 31. Genetics. 2011. PMID: 21196524 Free PMC article.
-
Host-Specific and Segment-Specific Evolutionary Dynamics of Avian and Human Influenza A Viruses: A Systematic Review.PLoS One. 2016 Jan 13;11(1):e0147021. doi: 10.1371/journal.pone.0147021. eCollection 2016. PLoS One. 2016. PMID: 26760775 Free PMC article. Review.
Cited by
-
Genetic Variability Under the Seedbank Coalescent.Genetics. 2015 Jul;200(3):921-34. doi: 10.1534/genetics.115.176818. Epub 2015 May 7. Genetics. 2015. PMID: 25953769 Free PMC article.
-
Investigation of the demographic and selective forces shaping the nucleotide diversity of genes involved in nod factor signaling in Medicago truncatula.Genetics. 2007 Dec;177(4):2123-33. doi: 10.1534/genetics.107.076943. Genetics. 2007. PMID: 18073426 Free PMC article.
-
Sequence diversity of Pan troglodytes subspecies and the impact of WFDC6 selective constraints in reproductive immunity.Genome Biol Evol. 2013;5(12):2512-23. doi: 10.1093/gbe/evt198. Genome Biol Evol. 2013. PMID: 24356879 Free PMC article.
-
Power of neutrality tests for detecting natural selection.G3 (Bethesda). 2023 Sep 30;13(10):jkad161. doi: 10.1093/g3journal/jkad161. G3 (Bethesda). 2023. PMID: 37481468 Free PMC article.
-
Detecting selection in the blue crab, Callinectes sapidus, using DNA sequence data from multiple nuclear protein-coding genes.PLoS One. 2014 Jun 4;9(6):e99081. doi: 10.1371/journal.pone.0099081. eCollection 2014. PLoS One. 2014. PMID: 24896825 Free PMC article.
References
-
- Bustamante, C. D., R. Nielsen, S. A. Sawyer, K. M. Olsen, M. D. Purugganan et al., 2002. The cost of inbreeding in Arabidopsis. Nature 416: 531–534. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

