Development of macrophages with altered actin organization in the absence of MafB
- PMID: 16943423
- PMCID: PMC1592864
- DOI: 10.1128/MCB.00245-06
Development of macrophages with altered actin organization in the absence of MafB
Abstract
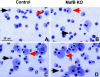
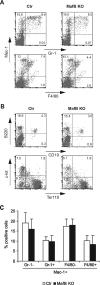
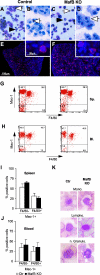
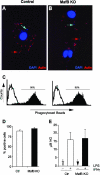
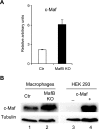
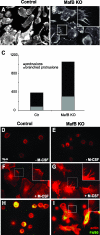
In the hematopoietic system the bZip transcription factor MafB is selectively expressed at high levels in monocytes and macrophages and promotes macrophage differentiation in myeloid progenitors, whereas a dominant-negative allele can inhibit this process. To analyze the requirement of MafB for macrophage development, we generated MafB-deficient mice and, due to their neonatal lethal phenotype, analyzed macrophage differentiation in vitro, in the embryo, and in reconstituted mice. Surprisingly we observed in vitro differentiation of macrophages from E14.5 fetal liver (FL) cells and E18.5 splenocytes. Furthermore we found normal numbers of F4/80(+)/Mac-1(+) macrophages and monocytes in fetal liver, spleen, and blood as well as in bone marrow, spleen, and peritoneum of adult MafB(-/-) FL reconstituted mice. MafB(-/-) macrophages showed intact basic macrophage functions such as phagocytosis of latex beads or Listeria monocytogenes and nitric oxide production in response to lipopolysaccharide. By contrast, MafB(-/-) macrophages expressed increased levels of multiple genes involved in actin organization. Consistent with this, phalloidin staining revealed an altered morphology involving increased numbers of branched protrusions of MafB(-/-) macrophages in response to macrophage colony-stimulating factor. Together these data point to an unexpected redundancy of MafB function in macrophage differentiation and a previously unknown role in actin-dependent macrophage morphology.
Figures


Similar articles
-
MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages.Science. 2009 Nov 6;326(5954):867-71. doi: 10.1126/science.1176056. Science. 2009. PMID: 19892988
-
SUMO modification regulates MafB-driven macrophage differentiation by enabling Myb-dependent transcriptional repression.Mol Cell Biol. 2007 Aug;27(15):5554-64. doi: 10.1128/MCB.01811-06. Epub 2007 Jun 4. Mol Cell Biol. 2007. PMID: 17548468 Free PMC article.
-
Differential expression patterns of MafB and c-Maf in macrophages in vivo and in vitro.Biochem Biophys Res Commun. 2016 Apr 22;473(1):118-124. doi: 10.1016/j.bbrc.2016.03.063. Epub 2016 Mar 17. Biochem Biophys Res Commun. 2016. PMID: 26996125
-
Development, differentiation, and maturation of Kupffer cells.Microsc Res Tech. 1997 Nov 15;39(4):350-64. doi: 10.1002/(SICI)1097-0029(19971115)39:4<350::AID-JEMT5>3.0.CO;2-L. Microsc Res Tech. 1997. PMID: 9407545 Review.
-
Role of MafB in macrophages.Exp Anim. 2020 Jan 29;69(1):1-10. doi: 10.1538/expanim.19-0076. Epub 2019 Oct 1. Exp Anim. 2020. PMID: 31582643 Free PMC article. Review.
Cited by
-
In Vitro Control of Genes Critical for Parathyroid Embryogenesis by Extracellular Calcium.J Endocr Soc. 2020 May 25;4(7):bvaa058. doi: 10.1210/jendso/bvaa058. eCollection 2020 Jul 1. J Endocr Soc. 2020. PMID: 32666007 Free PMC article.
-
Exploring Large MAF Transcription Factors: Functions, Pathology, and Mouse Models with Point Mutations.Genes (Basel). 2023 Sep 27;14(10):1883. doi: 10.3390/genes14101883. Genes (Basel). 2023. PMID: 37895232 Free PMC article. Review.
-
Role of large MAF transcription factors in the mouse endocrine pancreas.Exp Anim. 2015;64(3):305-12. doi: 10.1538/expanim.15-0001. Epub 2015 Apr 27. Exp Anim. 2015. PMID: 25912440 Free PMC article.
-
MafB is a critical regulator of complement component C1q.Nat Commun. 2017 Nov 22;8(1):1700. doi: 10.1038/s41467-017-01711-0. Nat Commun. 2017. PMID: 29167450 Free PMC article.
-
Role of H2-calponin in regulating macrophage motility and phagocytosis.J Biol Chem. 2008 Sep 19;283(38):25887-99. doi: 10.1074/jbc.M801163200. Epub 2008 Jul 9. J Biol Chem. 2008. PMID: 18617524 Free PMC article.
References
-
- Aggarwal, B. B., and K. Mehta. 1996. Determination and regulation of nitric oxide production from macrophages by lipopolysaccharides, cytokines, and retinoids. Methods Enzymol. 269:166-171. - PubMed
-
- Allen, W. E., G. E. Jones, J. W. Pollard, and A. J. Ridley. 1997. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110:707-720. - PubMed
-
- Bakri, Y., S. Sarrazin, U. P. Mayer, S. Tillmanns, C. Nerlov, A. Boned, and M. H. Sieweke. 2005. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 105:2707-2716. - PubMed
-
- Blanchi, B., L. M. Kelly, J. C. Viemari, I. Lafon, H. Burnet, M. Bevengut, S. Tillmanns, L. Daniel, T. Graf, G. Hilaire, and M. H. Sieweke. 2003. MafB deficiency causes defective respiratory rhythmogenesis and fatal central apnea at birth. Nat. Neurosci. 6:1091-1100. - PubMed
-
- Blanchi, B., and M. H. Sieweke. 2005. Mutations of brainstem transcription factors and central respiratory disorders. Trends Mol. Med. 11:23-30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
