NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila
- PMID: 16940169
- PMCID: PMC2118400
- DOI: 10.1084/jem.20060766
NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila
Abstract
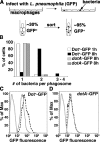
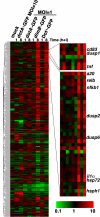
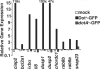
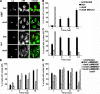
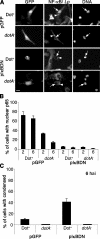
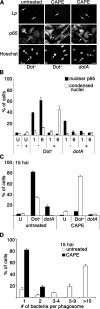
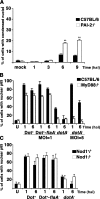
Legionella pneumophila, the causative agent of Legionnaires' disease, grows within macrophages and manipulates target cell signaling. Formation of a Legionella-containing replication vacuole requires the function of the bacterial type IV secretion system (Dot/Icm), which transfers protein substrates into the host cell cytoplasm. A global microarray analysis was used to examine the response of human macrophage-like U937 cells to low-dose infections with L. pneumophila. The most striking change in expression was the Dot/Icm-dependent up-regulation of antiapoptotic genes positively controlled by the transcriptional regulator nuclear factor kappaB (NF-kappaB). Consistent with this finding, L. pneumophila triggered nuclear localization of NF-kappaB in human and mouse macrophages in a Dot/Icm-dependent manner. The mechanism of activation at low-dose infections involved a signaling pathway that occurred independently of the Toll-like receptor adaptor MyD88 and the cytoplasmic sensor Nod1. In contrast, high multiplicity of infection conditions caused a host cell response that masked the unique Dot/Icm-dependent activation of NF-kappaB. Inhibition of NF-kappaB translocation into the nucleus resulted in premature host cell death and termination of bacterial replication. In the absence of one antiapoptotic protein, plasminogen activator inhibitor-2, host cell death increased in response to L. pneumophila infection, indicating that induction of antiapoptotic genes is critical for host cell survival.
Figures
Similar articles
-
Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila.Cell Microbiol. 2007 Jan;9(1):246-64. doi: 10.1111/j.1462-5822.2006.00785.x. Epub 2006 Aug 15. Cell Microbiol. 2007. PMID: 16911566
-
Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila.PLoS Pathog. 2011 Feb;7(2):e1001289. doi: 10.1371/journal.ppat.1001289. Epub 2011 Feb 17. PLoS Pathog. 2011. PMID: 21390206 Free PMC article.
-
MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires' disease.Infect Immun. 2006 Jun;74(6):3325-33. doi: 10.1128/IAI.02049-05. Infect Immun. 2006. PMID: 16714560 Free PMC article.
-
Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors.Cell Microbiol. 2011 Dec;13(12):1870-80. doi: 10.1111/j.1462-5822.2011.01710.x. Epub 2011 Nov 10. Cell Microbiol. 2011. PMID: 21981078 Review.
-
Cell biology of Legionella pneumophila.Curr Opin Microbiol. 1999 Feb;2(1):30-4. doi: 10.1016/s1369-5274(99)80005-8. Curr Opin Microbiol. 1999. PMID: 10047559 Review.
Cited by
-
Genetic susceptibility and caspase activation in mouse and human macrophages are distinct for Legionella longbeachae and L. pneumophila.Infect Immun. 2007 Apr;75(4):1933-45. doi: 10.1128/IAI.00025-07. Epub 2007 Jan 29. Infect Immun. 2007. PMID: 17261610 Free PMC article.
-
The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA.Cell Host Microbe. 2009 Sep 17;6(3):253-67. doi: 10.1016/j.chom.2009.08.005. Cell Host Microbe. 2009. PMID: 19748467 Free PMC article.
-
Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila.PLoS Pathog. 2008 Nov;4(11):e1000220. doi: 10.1371/journal.ppat.1000220. Epub 2008 Nov 28. PLoS Pathog. 2008. PMID: 19043549 Free PMC article.
-
The TLR4 D299G and T399I SNPs are constitutively active to up-regulate expression of Trif-dependent genes.PLoS One. 2014 Nov 3;9(11):e111460. doi: 10.1371/journal.pone.0111460. eCollection 2014. PLoS One. 2014. PMID: 25365308 Free PMC article.
-
Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella.Trends Microbiol. 2012 Jun;20(6):299-306. doi: 10.1016/j.tim.2012.03.005. Epub 2012 Apr 9. Trends Microbiol. 2012. PMID: 22494803 Free PMC article. Review.
References
-
- Fraser, D.W., T.R. Tsai, W. Orenstein, W.E. Parkin, H.J. Beecham, R.G. Sharrar, J. Harris, G.F. Mallison, S.M. Martin, J.E. McDade, et al. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189–1197. - PubMed
-
- Fields, B.S. 1996. The molecular ecology of Legionellae. Trends Microbiol. 4:286–290. - PubMed
-
- Tilney, L.G., O.S. Harb, P.S. Connelly, C.G. Robinson, and C.R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637–4650. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

