Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila
- PMID: 16938835
- PMCID: PMC1569195
- DOI: 10.1073/pnas.0604661103
Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila
Abstract
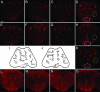
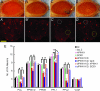
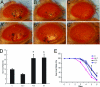
Parkinson's disease (PD) is the most frequent neurodegenerative movement disorder. Mutations in the PINK1 gene are linked to the autosomal recessive early onset familial form of PD. The physiological function of PINK1 and pathological abnormality of PD-associated PINK1 mutants are largely unknown. We here show that inactivation of Drosophila PINK1 (dPINK1) using RNAi results in progressive loss of dopaminergic neurons and in ommatidial degeneration of the compound eye, which is rescued by expression of human PINK1 (hPINK1). Expression of human SOD1 suppresses neurodegeneration induced by dPINK1 inactivation. Moreover, treatment of dPINK1 RNAi flies with the antioxidants SOD and vitamin E significantly inhibits ommatidial degeneration. Thus, dPINK1 plays an essential role in maintaining neuronal survival by preventing neurons from undergoing oxidative stress, thereby suggesting a potential mechanism by which a reduction in PINK1 function leads to PD-associated neurodegeneration.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

Comment in
-
Antioxidants put Parkinson flies back in the PINK.Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13269-70. doi: 10.1073/pnas.0606288103. Epub 2006 Aug 28. Proc Natl Acad Sci U S A. 2006. PMID: 16938856 Free PMC article. No abstract available.
Similar articles
-
Antioxidants put Parkinson flies back in the PINK.Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13269-70. doi: 10.1073/pnas.0606288103. Epub 2006 Aug 28. Proc Natl Acad Sci U S A. 2006. PMID: 16938856 Free PMC article. No abstract available.
-
Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin.Proc Natl Acad Sci U S A. 2006 Jul 11;103(28):10793-8. doi: 10.1073/pnas.0602493103. Epub 2006 Jul 3. Proc Natl Acad Sci U S A. 2006. PMID: 16818890 Free PMC article.
-
Pink1 suppresses alpha-synuclein-induced phenotypes in a Drosophila model of Parkinson's disease.Genome. 2008 Dec;51(12):1040-6. doi: 10.1139/G08-085. Genome. 2008. PMID: 19088817
-
Genetic mutations and functions of PINK1.Trends Pharmacol Sci. 2011 Oct;32(10):573-80. doi: 10.1016/j.tips.2011.06.001. Epub 2011 Jul 23. Trends Pharmacol Sci. 2011. PMID: 21784538 Review.
-
PINK1 mediates neuronal survival in monkey.Protein Cell. 2022 Jan;13(1):4-5. doi: 10.1007/s13238-021-00889-w. Epub 2021 Nov 23. Protein Cell. 2022. PMID: 34813054 Free PMC article. Review. No abstract available.
Cited by
-
PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1.PLoS Biol. 2007 Jul;5(7):e172. doi: 10.1371/journal.pbio.0050172. Epub 2007 Jun 19. PLoS Biol. 2007. PMID: 17579517 Free PMC article.
-
Neurodegenerative models in Drosophila: polyglutamine disorders, Parkinson disease, and amyotrophic lateral sclerosis.Neurobiol Dis. 2010 Oct;40(1):29-39. doi: 10.1016/j.nbd.2010.05.026. Epub 2010 May 31. Neurobiol Dis. 2010. PMID: 20561920 Free PMC article. Review.
-
Drosophila models of Parkinson's disease: discovering relevant pathways and novel therapeutic strategies.Parkinsons Dis. 2011 Mar 3;2011:520640. doi: 10.4061/2011/520640. Parkinsons Dis. 2011. PMID: 21512585 Free PMC article.
-
FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation.Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):5153-8. doi: 10.1073/pnas.0901104106. Epub 2009 Mar 10. Proc Natl Acad Sci U S A. 2009. PMID: 19276113 Free PMC article.
-
Developmental nicotine exposure affects larval brain size and the adult dopaminergic system of Drosophila melanogaster.BMC Dev Biol. 2018 Jun 14;18(1):13. doi: 10.1186/s12861-018-0172-6. BMC Dev Biol. 2018. PMID: 29898654 Free PMC article.
References
-
- Lang A. E., Lozano A. M. N. Engl. J. Med. 1998;339:1130–1143. - PubMed
-
- Lang A. E., Lozano A. M. N. Engl. J. Med. 1998;339:1044–1053. - PubMed
-
- Dawson T. M., Dawson V. L. Science. 2003;302:819–822. - PubMed
-
- Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., et al. Science. 2004;304:1158–1160. - PubMed
-
- Paisan-Ruiz C., Jain S., Evans E. W., Gilks W. P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A. M., Khan N., et al. Neuron. 2004;44:595–600. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

