CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding
- PMID: 16938098
- PMCID: PMC1652823
- DOI: 10.1042/BJ20060490
CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding
Abstract
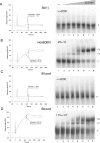
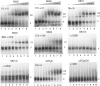
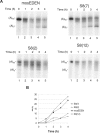
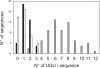
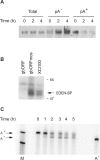
CUG-BP1 [CUG-binding protein 1 also called CELF (CUG-BP1 and ETR3 like factors) 1] is a human RNA-binding protein that has been implicated in the control of splicing and mRNA translation. The Xenopus homologue [EDEN-BP (embryo deadenylation element-binding protein)] is required for rapid deadenylation of certain maternal mRNAs just after fertilization. A variety of sequence elements have been described as target sites for these two proteins but their binding specificity is still controversial. Using a SELEX (systematic evolution of ligand by exponential enrichment) procedure and recombinant CUG-BP1 we selected two families of aptamers. Surface plasmon resonance and electrophoretic mobility-shift assays showed that these two families differed in their ability to bind CUG-BP1. Furthermore, the selected high-affinity aptamers form two complexes with CUG-BP1 in electrophoretic mobility assays whereas those that bind with low affinity only form one complex. The validity of the distinction between the two families of aptamers was confirmed by a functional in vivo deadenylation assay. Only those aptamers that bound CUG-BP1 with high affinity conferred deadenylation on a reporter mRNA. These high-affinity RNAs are characterized by a richness in UGU motifs. Using these binding site characteristics we identified the Xenopus maternal mRNA encoding the MAPK (mitogen-activated protein kinase) phosphatase (XCl100alpha) as a substrate for EDEN-BP. In conclusion, high-affinity CUG-BP1 binding sites are sequence elements at least 30 nucleotides in length that are enriched in combinations of U and G nucleotides and contain at least 4 UGU trinucleotide motifs. Such sequence elements are functionally competent to target an RNA for deadenylation in vivo.
Figures
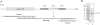
Similar articles
-
Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1.Curr Opin Genet Dev. 2011 Aug;21(4):444-51. doi: 10.1016/j.gde.2011.03.002. Epub 2011 Apr 13. Curr Opin Genet Dev. 2011. PMID: 21497082 Free PMC article. Review.
-
Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis.Nucleic Acids Res. 2008 Apr;36(6):1861-70. doi: 10.1093/nar/gkn031. Epub 2008 Feb 11. Nucleic Acids Res. 2008. PMID: 18267972 Free PMC article.
-
c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway.J Biol Chem. 2002 Feb 1;277(5):3232-5. doi: 10.1074/jbc.M109362200. Epub 2001 Nov 13. J Biol Chem. 2002. PMID: 11707455
-
A functional deadenylation assay identifies human CUG-BP as a deadenylation factor.Biol Cell. 2003 Mar-Apr;95(2):107-13. doi: 10.1016/s0248-4900(03)00010-8. Biol Cell. 2003. PMID: 12799066
-
East of EDEN was a poly(A) tail.Biol Cell. 2003 May-Jun;95(3-4):211-9. doi: 10.1016/s0248-4900(03)00038-8. Biol Cell. 2003. PMID: 12867084 Review.
Cited by
-
A protein assembly mediates Xist localization and gene silencing.Nature. 2020 Nov;587(7832):145-151. doi: 10.1038/s41586-020-2703-0. Epub 2020 Sep 9. Nature. 2020. PMID: 32908311 Free PMC article.
-
High-throughput binding analysis determines the binding specificity of ASF/SF2 on alternatively spliced human pre-mRNAs.Comb Chem High Throughput Screen. 2010 Mar;13(3):242-52. doi: 10.2174/138620710790980522. Comb Chem High Throughput Screen. 2010. PMID: 20015017 Free PMC article.
-
miR-322 promotes the differentiation of embryonic stem cells into cardiomyocytes.Funct Integr Genomics. 2023 Mar 18;23(2):87. doi: 10.1007/s10142-023-01008-0. Funct Integr Genomics. 2023. PMID: 36932296
-
Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells.PLoS One. 2010 Jun 21;5(6):e11201. doi: 10.1371/journal.pone.0011201. PLoS One. 2010. PMID: 20574513 Free PMC article.
-
Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1.Curr Opin Genet Dev. 2011 Aug;21(4):444-51. doi: 10.1016/j.gde.2011.03.002. Epub 2011 Apr 13. Curr Opin Genet Dev. 2011. PMID: 21497082 Free PMC article. Review.
References
-
- Timchenko L. T., Timchenko N. A., Caskey C. T., Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet. 1996;5:115–121. - PubMed
-
- Barreau C., Paillard L., Mereau A., Osborne H. B. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. - PubMed
-
- Philips A. V., Timchenko L. T., Cooper T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. - PubMed
-
- Ho T. H., Bundman D., Armstrong D. L., Cooper T. A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

