Structural basis of ubiquitin recognition by the deubiquitinating protease USP2
- PMID: 16905103
- PMCID: PMC7126176
- DOI: 10.1016/j.str.2006.06.012
Structural basis of ubiquitin recognition by the deubiquitinating protease USP2
Abstract
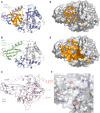
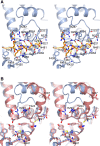
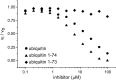
Deubiquitinating proteases reverse protein ubiquitination and rescue their target proteins from destruction by the proteasome. USP2, a cysteine protease and a member of the ubiquitin specific protease family, is overexpressed in prostate cancer and stabilizes fatty acid synthase, which has been associated with the malignancy of some aggressive prostate cancers. Here, we report the structure of the human USP2 catalytic domain in complex with ubiquitin. Ubiquitin uses two major sites for the interaction with the protease. Both sites are required simultaneously, as shown by USP2 inhibition assays with peptides and ubiquitin mutants. In addition, a layer of ordered water molecules mediates key interactions between ubiquitin and USP2. As several of those molecules are found at identical positions in the previously solved USP7/ubiquitin-aldehyde complex structure, we suggest a general mechanism of water-mediated ubiquitin recognition by USPs.
Figures

Similar articles
-
Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases.Biochemistry. 2011 May 31;50(21):4775-85. doi: 10.1021/bi101958h. Epub 2011 May 4. Biochemistry. 2011. PMID: 21542621
-
Validation of catalytic site residues of Ubiquitin Specific Protease 2 (USP2) by molecular dynamic simulation and novel kinetics assay for rational drug design.Mol Divers. 2023 Jun;27(3):1323-1332. doi: 10.1007/s11030-022-10499-1. Epub 2022 Aug 6. Mol Divers. 2023. PMID: 35932436
-
Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp2-45 and Usp2-69.Gene Expr. 2003;11(3-4):163-79. doi: 10.3727/000000003108749053. Gene Expr. 2003. PMID: 14686789 Free PMC article.
-
The role of UBL domains in ubiquitin-specific proteases.Biochem Soc Trans. 2012 Jun 1;40(3):539-45. doi: 10.1042/BST20120004. Biochem Soc Trans. 2012. PMID: 22616864 Review.
-
Ubiquitin-Specific Proteases (USPs) and Metabolic Disorders.Int J Mol Sci. 2023 Feb 6;24(4):3219. doi: 10.3390/ijms24043219. Int J Mol Sci. 2023. PMID: 36834633 Free PMC article. Review.
Cited by
-
Ubiquitylation of nucleic acids by DELTEX ubiquitin E3 ligase DTX3L.EMBO Rep. 2024 Oct;25(10):4172-4189. doi: 10.1038/s44319-024-00235-1. Epub 2024 Sep 6. EMBO Rep. 2024. PMID: 39242775 Free PMC article.
-
Deubiquitinating enzymes as novel anticancer targets.Future Oncol. 2007 Apr;3(2):191-9. doi: 10.2217/14796694.3.2.191. Future Oncol. 2007. PMID: 17381419 Free PMC article. Review.
-
The pathogenesis of prostate cancer: from molecular to metabolic alterations.Diagn Histopathol (Oxf). 2008 May;14(5):195-201. doi: 10.1016/j.mpdhp.2008.03.001. Diagn Histopathol (Oxf). 2008. PMID: 20953243 Free PMC article.
-
De novo macrocyclic peptides that specifically modulate Lys48-linked ubiquitin chains.Nat Chem. 2019 Jul;11(7):644-652. doi: 10.1038/s41557-019-0278-x. Epub 2019 Jun 10. Nat Chem. 2019. PMID: 31182821 Free PMC article.
-
Downregulation of ubiquitin-specific protease 2 possesses prognostic and diagnostic value and promotes the clear cell renal cell carcinoma progression.Ann Transl Med. 2020 Mar;8(6):319. doi: 10.21037/atm.2020.02.141. Ann Transl Med. 2020. PMID: 32355763 Free PMC article.
References
-
- Abergel C. Spectacular improvement of X-ray diffraction through fast desiccation of protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1413–1416. - PubMed
-
- Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. - PubMed
-
- Baron A., Migita T., Tang D., Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J. Cell. Biochem. 2004;91:47–53. - PubMed
-
- Cummins J.M., Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

