STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments
- PMID: 16899652
- PMCID: PMC1557772
- DOI: 10.1101/gr.5076506
STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments
Abstract
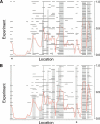
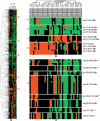
Regions of gain and loss of genomic DNA occur in many cancers and can drive the genesis and progression of disease. These copy number aberrations (CNAs) can be detected at high resolution by using microarray-based techniques. However, robust statistical approaches are needed to identify nonrandom gains and losses across multiple experiments/samples. We have developed a method called Significance Testing for Aberrant Copy number (STAC) to address this need. STAC utilizes two complementary statistics in combination with a novel search strategy. The significance of both statistics is assessed, and P-values are assigned to each location on the genome by using a multiple testing corrected permutation approach. We validate our method by using two published cancer data sets. STAC identifies genomic alterations known to be of clinical and biological significance and provides statistical support for 85% of previously reported regions. Moreover, STAC identifies numerous additional regions of significant gain/loss in these data that warrant further investigation. The P-values provided by STAC can be used to prioritize regions for follow-up study in an unbiased fashion. We conclude that STAC is a powerful tool for identifying nonrandom genomic amplifications and deletions across multiple experiments. A Java version of STAC is freely available for download at http://cbil.upenn.edu/STAC.
Figures



Similar articles
-
Assessing the significance of conserved genomic aberrations using high resolution genomic microarrays.PLoS Genet. 2007 Aug;3(8):e143. doi: 10.1371/journal.pgen.0030143. PLoS Genet. 2007. PMID: 17722985 Free PMC article.
-
Functionally-focused algorithmic analysis of high resolution microarray-CGH genomic landscapes demonstrates comparable genomic copy number aberrations in MSI and MSS sporadic colorectal cancer.PLoS One. 2017 Feb 23;12(2):e0171690. doi: 10.1371/journal.pone.0171690. eCollection 2017. PLoS One. 2017. PMID: 28231327 Free PMC article.
-
Accurate detection of aneuploidies in array CGH and gene expression microarray data.Bioinformatics. 2004 Dec 12;20(18):3533-43. doi: 10.1093/bioinformatics/bth440. Epub 2004 Jul 29. Bioinformatics. 2004. PMID: 15284100
-
Current status and future prospects of array-based comparative genomic hybridisation.Brief Funct Genomic Proteomic. 2003 Apr;2(1):37-45. doi: 10.1093/bfgp/2.1.37. Brief Funct Genomic Proteomic. 2003. PMID: 15239942 Review.
-
Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors.Genes Chromosomes Cancer. 1994 Aug;10(4):231-43. doi: 10.1002/gcc.2870100403. Genes Chromosomes Cancer. 1994. PMID: 7522536 Review.
Cited by
-
A scale-space method for detecting recurrent DNA copy number changes with analytical false discovery rate control.Nucleic Acids Res. 2013 May;41(9):e100. doi: 10.1093/nar/gkt155. Epub 2013 Mar 8. Nucleic Acids Res. 2013. PMID: 23476020 Free PMC article.
-
GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers.Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. Epub 2011 Apr 28. Genome Biol. 2011. PMID: 21527027 Free PMC article.
-
Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia.Haematologica. 2011 Aug;96(8):1161-9. doi: 10.3324/haematol.2010.039768. Epub 2011 May 5. Haematologica. 2011. PMID: 21546498 Free PMC article.
-
CGHweb: a tool for comparing DNA copy number segmentations from multiple algorithms.Bioinformatics. 2008 Apr 1;24(7):1014-5. doi: 10.1093/bioinformatics/btn067. Epub 2008 Feb 22. Bioinformatics. 2008. PMID: 18296463 Free PMC article.
-
Mutational Analysis of Ionizing Radiation Induced Neoplasms.Cell Rep. 2015 Sep 22;12(11):1915-26. doi: 10.1016/j.celrep.2015.08.015. Epub 2015 Sep 3. Cell Rep. 2015. PMID: 26344771 Free PMC article.
References
-
- Aguirre A.J., Brennan C., Baily G., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Brennan C., Baily G., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Baily G., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Feng B., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Leo C., Zhang Y., Zhang J., Gans J.D., Bardessy N., Zhang Y., Zhang J., Gans J.D., Bardessy N., Zhang J., Gans J.D., Bardessy N., Gans J.D., Bardessy N., Bardessy N., et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc. Natl. Acad. Sci. 2004;101:9067–9072. - PMC - PubMed
-
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Adams M.D., Meyers E.W., Li P.W., Eichler E.E., Meyers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Barrett M.T., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Tsang P., Curry B., Baird K., Meltzer P.S., Curry B., Baird K., Meltzer P.S., Baird K., Meltzer P.S., Meltzer P.S., et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl. Acad. Sci. 2004;101:17765–17770. - PMC - PubMed
-
- Brodeur G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3:203–216. - PubMed
-
- Brodeur G.M., Maris J.M., Maris J.M. In: Principles and practice of pediatric oncology. 4th ed. Pizzo P.A., Pollack D.G., Pollack D.G., editors. Lippincott Williams & Wilkins; Philadelphia PA: 2002. pp. 895–938.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
