Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana
- PMID: 16897492
- PMCID: PMC3574581
- DOI: 10.1007/s11103-006-0049-0
Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana
Abstract
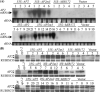
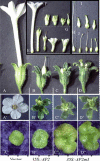
Floral patterning and morphogenesis are controlled by many transcription factors including floral homeotic proteins, by which floral organ identity is determined. Recent studies have uncovered widespread regulation of transcription factors by microRNAs (miRNAs), approximately 21-nucleotide non-coding RNAs that regulate protein-coding RNAs through transcript cleavage and/or translational inhibition. The regulation of the floral homeotic gene APETALA2 (AP2) by miR172 is crucial for normal Arabidopsis flower development and is likely to be conserved across plant species. Here we probe the activity of the AP2/miR172 regulatory circuit in a heterologous Solanaceae species, Nicotiana benthamiana. We generated transgenic N. benthamiana lines expressing Arabidopsis wild type AP2 (35S::AP2), miR172-resistant AP2 mutant (35S::AP2m3) and MIR172a-1 (35S::MIR172) under the control of the cauliflower mosaic virus 35S promoter. 35S::AP2m3 plants accumulated high levels of AP2 mRNA and protein and exhibited floral patterning defects that included proliferation of numerous petals, stamens and carpels indicating loss of floral determinacy. On the other hand, nearly all 35S::AP2 plants accumulated barely detectable levels of AP2 mRNA or protein and were essentially non-phenotypic. Overall, the data indicated that expression of the wild type Arabidopsis AP2 transgene was repressed at the mRNA level by an endogenous N. benthamiana miR172 homologue that could be detected using Arabidopsis miR172 probe. Interestingly, 35S::MIR172 plants had sepal-to-petal transformations and/or more sepals and petals, suggesting interference with N. benthamiana normal floral homeotic gene function in perianth organs. Our studies uncover the potential utility of the Arabidopsis AP2/miR172 system as a tool for manipulation of floral architecture and flowering time in non-model plants.
Figures




Similar articles
-
The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning.Plant Sci. 2014 Feb;215-216:29-38. doi: 10.1016/j.plantsci.2013.10.010. Epub 2013 Oct 25. Plant Sci. 2014. PMID: 24388512
-
On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development.Development. 2010 Nov;137(21):3633-42. doi: 10.1242/dev.036673. Epub 2010 Sep 28. Development. 2010. PMID: 20876650 Free PMC article.
-
miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems.Plant J. 2007 Sep;51(5):840-9. doi: 10.1111/j.1365-313X.2007.03181.x. Epub 2007 Jun 15. Plant J. 2007. PMID: 17573799 Free PMC article.
-
Regulation of flowering time and floral patterning by miR172.J Exp Bot. 2011 Jan;62(2):487-95. doi: 10.1093/jxb/erq295. Epub 2010 Oct 15. J Exp Bot. 2011. PMID: 20952628 Review.
-
Sculpting the flower; the role of microRNAs in flower development.Curr Top Dev Biol. 2010;91:349-78. doi: 10.1016/S0070-2153(10)91012-0. Curr Top Dev Biol. 2010. PMID: 20705188 Review.
Cited by
-
The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element.Development. 2012 Jun;139(11):1978-86. doi: 10.1242/dev.077073. Epub 2012 Apr 18. Development. 2012. PMID: 22513376 Free PMC article.
-
TM8 represses developmental timing in Nicotiana benthamiana and has functionally diversified in angiosperms.BMC Plant Biol. 2018 Jun 22;18(1):129. doi: 10.1186/s12870-018-1349-7. BMC Plant Biol. 2018. PMID: 29929474 Free PMC article.
-
Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress.BMC Plant Biol. 2016 Jul 30;16(1):171. doi: 10.1186/s12870-016-0859-4. BMC Plant Biol. 2016. PMID: 27475988 Free PMC article.
-
The Roles of Floral Organ Genes in Regulating Rosaceae Fruit Development.Front Plant Sci. 2022 Jan 5;12:644424. doi: 10.3389/fpls.2021.644424. eCollection 2021. Front Plant Sci. 2022. PMID: 35069608 Free PMC article. Review.
-
The APETALA2 homolog CaFFN regulates flowering time in pepper.Hortic Res. 2021 Nov 1;8(1):208. doi: 10.1038/s41438-021-00643-7. Hortic Res. 2021. PMID: 34719686 Free PMC article.
References
-
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;31:3357–3365. - PubMed
-
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

