Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release
- PMID: 16894145
- PMCID: PMC1567914
- DOI: 10.1073/pnas.0605402103
Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release
Abstract
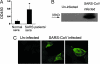
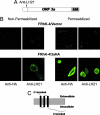
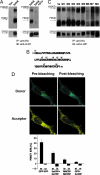
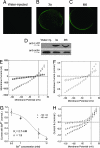
Fourteen ORFs have been identified in the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) genome. ORF 3a of SARS-CoV codes for a recently identified transmembrane protein, but its function remains unknown. In this study we confirmed the 3a protein expression and investigated its localization at the surface of SARS-CoV-infected or 3a-cDNA-transfected cells. Our experiments showed that recombinant 3a protein can form a homotetramer complex through interprotein disulfide bridges in 3a-cDNA-transfected cells, providing a clue to ion channel function. The putative ion channel activity of this protein was assessed in 3a-complement RNA-injected Xenopus oocytes by two-electrode voltage clamp. The results suggest that 3a protein forms a potassium sensitive channel, which can be efficiently inhibited by barium. After FRhK-4 cells were transfected with an siRNA, which is known to suppress 3a expression, followed by infection with SARS-CoV, the released virus was significantly decreased, whereas the replication of the virus in the infected cells was not changed. Our observation suggests that SARS-CoV ORF 3a functions as an ion channel that may promote virus release. This finding will help to explain the highly pathogenic nature of SARS-CoV and to develop new strategies for treatment of SARS infection.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Similar articles
-
The Severe Acute Respiratory Syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein.Virol J. 2005 Feb 10;2:5. doi: 10.1186/1743-422X-2-5. Virol J. 2005. PMID: 15703085 Free PMC article.
-
Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis.mBio. 2018 May 22;9(3):e02325-17. doi: 10.1128/mBio.02325-17. mBio. 2018. PMID: 29789363 Free PMC article.
-
The SARS Coronavirus 3a protein binds calcium in its cytoplasmic domain.Virus Res. 2014 Oct 13;191:180-3. doi: 10.1016/j.virusres.2014.08.001. Epub 2014 Aug 10. Virus Res. 2014. PMID: 25116391 Free PMC article.
-
Coronavirus Proteins as Ion Channels: Current and Potential Research.Front Immunol. 2020 Oct 9;11:573339. doi: 10.3389/fimmu.2020.573339. eCollection 2020. Front Immunol. 2020. PMID: 33154751 Free PMC article. Review.
-
Expression of the severe acute respiratory syndrome coronavirus 3a protein and the assembly of coronavirus-like particles in the baculovirus expression system.Methods Mol Biol. 2007;379:35-50. doi: 10.1007/978-1-59745-393-6_3. Methods Mol Biol. 2007. PMID: 17502669 Free PMC article. Review.
Cited by
-
Severe acute respiratory syndrome coronavirus protein 6 mediates ubiquitin-dependent proteosomal degradation of N-Myc (and STAT) interactor.Virol Sin. 2015 Apr;30(2):153-61. doi: 10.1007/s12250-015-3581-8. Epub 2015 Apr 17. Virol Sin. 2015. PMID: 25907116 Free PMC article.
-
Molecular Evolution of Human Coronavirus Genomes.Trends Microbiol. 2017 Jan;25(1):35-48. doi: 10.1016/j.tim.2016.09.001. Epub 2016 Oct 19. Trends Microbiol. 2017. PMID: 27743750 Free PMC article. Review.
-
A novel diG motif in ORF3a protein of SARS-Cov-2 for intracellular transport.Front Cell Dev Biol. 2022 Nov 23;10:1011221. doi: 10.3389/fcell.2022.1011221. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506095 Free PMC article.
-
The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death.J Virol. 2010 Jan;84(2):1097-109. doi: 10.1128/JVI.01662-09. Epub 2009 Nov 4. J Virol. 2010. PMID: 19889773 Free PMC article.
-
Respiratory viruses and the inflammasome: The double-edged sword of inflammation.PLoS Pathog. 2022 Dec 29;18(12):e1011014. doi: 10.1371/journal.ppat.1011014. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36580480 Free PMC article. No abstract available.
References
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A., et al. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., et al. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Marra M. A., Jones S. J., Astell C. R., Holt R. A., Brooks-Wilson A., Butterfield Y. S., Khattra J., Asano J. K., Barber S. A., Chan S. Y., et al. Science. 2003;300:1399–1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

