Repair of topoisomerase I-mediated DNA damage
- PMID: 16891172
- PMCID: PMC2576451
- DOI: 10.1016/S0079-6603(06)81005-6
Repair of topoisomerase I-mediated DNA damage
Abstract
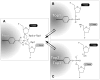
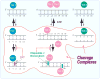
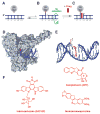
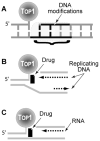
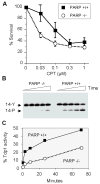
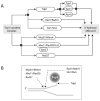
Topoisomerase I (Top1) is an abundant and essential enzyme. Top1 is the selective target of camptothecins, which are effective anticancer agents. Top1-DNA cleavage complexes can also be trapped by various endogenous and exogenous DNA lesions including mismatches, abasic sites and carcinogenic adducts. Tyrosyl-DNA phosphodiesterase (Tdp1) is one of the repair enzymes for Top1-DNA covalent complexes. Tdp1 forms a multiprotein complex that includes poly(ADP) ribose polymerase (PARP). PARP-deficient cells are hypersensitive to camptothecins and functionally deficient for Tdp1. We will review recent developments in several pathways involved in the repair of Top1 cleavage complexes and the role of Chk1 and Chk2 checkpoint kinases in the cellular responses to Top1 inhibitors. The genes conferring camptothecin hypersensitivity are compiled for humans, budding yeast and fission yeast.
Figures
Similar articles
-
DNA topoisomerases as targets for anticancer drugs.J Clin Pharm Ther. 2001 Dec;26(6):405-16. doi: 10.1046/j.1365-2710.2001.00368.x. J Clin Pharm Ther. 2001. PMID: 11722677
-
Symposium: downstream from topoisomerase-DNA lesions: life-or-death consequences for the cell.Cell Biochem Biophys. 2000;33(2):171-3. doi: 10.1385/CBB:33:2:171. Cell Biochem Biophys. 2000. PMID: 11325036 No abstract available.
-
Topoisomerase I-mediated DNA damage.Adv Cancer Res. 2001;80:189-216. doi: 10.1016/s0065-230x(01)80016-6. Adv Cancer Res. 2001. PMID: 11034544 Review.
-
Drug-induced stabilization of covalent DNA topoisomerase I-DNA intermediates. DNA cleavage assays.Methods Mol Biol. 2001;95:291-301. doi: 10.1385/1-59259-057-8:291. Methods Mol Biol. 2001. PMID: 11089241 No abstract available.
-
Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy.Biomolecules. 2015 Jul 22;5(3):1652-70. doi: 10.3390/biom5031652. Biomolecules. 2015. PMID: 26287259 Free PMC article. Review.
Cited by
-
Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast.DNA Repair (Amst). 2013 Mar 1;12(3):205-11. doi: 10.1016/j.dnarep.2012.12.004. Epub 2013 Jan 8. DNA Repair (Amst). 2013. PMID: 23305949 Free PMC article.
-
Potentiation of the novel topoisomerase I inhibitor indenoisoquinoline LMP-400 by the cell checkpoint and Chk1-Chk2 inhibitor AZD7762.Cancer Res. 2012 Feb 15;72(4):979-89. doi: 10.1158/0008-5472.CAN-11-2579. Epub 2011 Dec 21. Cancer Res. 2012. PMID: 22189968 Free PMC article.
-
Reduced expression of DNA topoisomerase I in SF295 human glioblastoma cells selected for resistance to homocamptothecin and diflomotecan.Mol Pharmacol. 2008 Feb;73(2):490-7. doi: 10.1124/mol.107.041178. Epub 2007 Nov 5. Mol Pharmacol. 2008. PMID: 17984197 Free PMC article.
-
Lignin Nanoparticles Deliver Novel Thymine Biomimetic Photo-Adducts with Antimelanoma Activity.Int J Mol Sci. 2022 Jan 14;23(2):915. doi: 10.3390/ijms23020915. Int J Mol Sci. 2022. PMID: 35055101 Free PMC article.
-
Combined inhibition of topoisomerase I and poly(ADP-ribose) polymerase: A synergistic therapeutic strategy for glioblastoma with phosphatase and tensin homolog deficiency.Neurooncol Adv. 2023 Aug 21;5(1):vdad102. doi: 10.1093/noajnl/vdad102. eCollection 2023 Jan-Dec. Neurooncol Adv. 2023. PMID: 37706203 Free PMC article.
References
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. - PubMed
-
- Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. - PubMed
-
- Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278:42668–42678. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

