The molecular biology of coronaviruses
- PMID: 16877062
- PMCID: PMC7112330
- DOI: 10.1016/S0065-3527(06)66005-3
The molecular biology of coronaviruses
Abstract
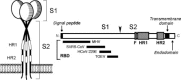
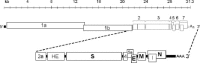
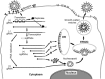
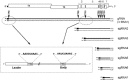
Coronaviruses are large, enveloped RNA viruses of both medical and veterinary importance. Interest in this viral family has intensified in the past few years as a result of the identification of a newly emerged coronavirus as the causative agent of severe acute respiratory syndrome (SARS). At the molecular level, coronaviruses employ a variety of unusual strategies to accomplish a complex program of gene expression. Coronavirus replication entails ribosome frameshifting during genome translation, the synthesis of both genomic and multiple subgenomic RNA species, and the assembly of progeny virions by a pathway that is unique among enveloped RNA viruses. Progress in the investigation of these processes has been enhanced by the development of reverse genetic systems, an advance that was heretofore obstructed by the enormous size of the coronavirus genome. This review summarizes both classical and contemporary discoveries in the study of the molecular biology of these infectious agents, with particular emphasis on the nature and recognition of viral receptors, viral RNA synthesis, and the molecular interactions governing virion assembly.
Figures


Similar articles
-
Coronaviruses: an overview of their replication and pathogenesis.Methods Mol Biol. 2015;1282:1-23. doi: 10.1007/978-1-4939-2438-7_1. Methods Mol Biol. 2015. PMID: 25720466 Free PMC article. Review.
-
Molecular mechanisms of coronavirus RNA capping and methylation.Virol Sin. 2016 Feb;31(1):3-11. doi: 10.1007/s12250-016-3726-4. Epub 2016 Feb 2. Virol Sin. 2016. PMID: 26847650 Free PMC article. Review.
-
Host Factors in Coronavirus Replication.Curr Top Microbiol Immunol. 2018;419:1-42. doi: 10.1007/82_2017_25. Curr Top Microbiol Immunol. 2018. PMID: 28643204 Free PMC article. Review.
-
Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus.Microbiol Mol Biol Rev. 2005 Dec;69(4):635-64. doi: 10.1128/MMBR.69.4.635-664.2005. Microbiol Mol Biol Rev. 2005. PMID: 16339739 Free PMC article. Review.
-
Viral and Cellular mRNA Translation in Coronavirus-Infected Cells.Adv Virus Res. 2016;96:165-192. doi: 10.1016/bs.aivir.2016.08.001. Epub 2016 Sep 10. Adv Virus Res. 2016. PMID: 27712623 Free PMC article. Review.
Cited by
-
SARS-CoV-2 vaccine research and development: Conventional vaccines and biomimetic nanotechnology strategies.Asian J Pharm Sci. 2021 Mar;16(2):136-146. doi: 10.1016/j.ajps.2020.08.001. Epub 2020 Sep 1. Asian J Pharm Sci. 2021. PMID: 32905011 Free PMC article. Review.
-
Potential 3-chymotrypsin-like cysteine protease cleavage sites in the coronavirus polyproteins pp1a and pp1ab and their possible relevance to COVID-19 vaccine and drug development.FASEB J. 2021 May;35(5):e21573. doi: 10.1096/fj.202100280RR. FASEB J. 2021. PMID: 33913206 Free PMC article. Review.
-
Animal Coronaviruses Induced Apoptosis.Life (Basel). 2021 Feb 26;11(3):185. doi: 10.3390/life11030185. Life (Basel). 2021. PMID: 33652685 Free PMC article. Review.
-
Reducing SARS-CoV-2 pathological protein activity with small molecules.J Pharm Anal. 2021 Aug;11(4):383-397. doi: 10.1016/j.jpha.2021.03.012. Epub 2021 Apr 5. J Pharm Anal. 2021. PMID: 33842018 Free PMC article. Review.
-
Mechanisms and implications of programmed translational frameshifting.Wiley Interdiscip Rev RNA. 2012 Sep-Oct;3(5):661-73. doi: 10.1002/wrna.1126. Epub 2012 Jun 19. Wiley Interdiscip Rev RNA. 2012. PMID: 22715123 Free PMC article. Review.
References
-
- Almeida J.D., Tyrrell D.A. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J. Gen. Virol. 1967;1:175–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

