Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10
- PMID: 16873247
- PMCID: PMC1563834
- DOI: 10.1128/JVI.00483-06
Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10
Abstract
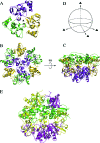
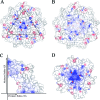
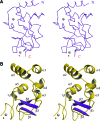
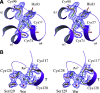
The severe acute respiratory syndrome coronavirus (SARS-CoV) nonstructural proteins nsp1 to nsp16 have been implicated by genetic analysis in the assembly of a functional replication/transcription complex. We report the crystal structure of nsp10 from SARS-CoV at 2.1-A resolution. The nsp10 structure has a novel fold, and 12 identical subunits assemble to form a unique spherical dodecameric architecture. Two zinc fingers have been identified from the nsp10 monomer structure with the sequence motifs C-(X)2-C-(X)5-H-(X)6-C and C-(X)2-C-(X)7-C-(X)-C. The nsp10 crystal structure is the first of a new class of zinc finger protein three-dimensional structures to be revealed experimentally. The zinc finger sequence motifs are conserved among all three coronavirus antigenic groups, implicating an essential function for nsp10 in all coronaviruses. Based on the structure, we propose that nsp10 is a transcription factor for coronavirus replication/transcription.
Figures

Similar articles
-
Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.J Virol. 2006 Aug;80(16):7894-901. doi: 10.1128/JVI.00467-06. J Virol. 2006. PMID: 16873246 Free PMC article.
-
Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.PLoS Pathog. 2011 Oct;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. Epub 2011 Oct 13. PLoS Pathog. 2011. PMID: 22022266 Free PMC article.
-
Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes.J Biol Chem. 2014 Sep 12;289(37):25783-96. doi: 10.1074/jbc.M114.577353. Epub 2014 Jul 29. J Biol Chem. 2014. PMID: 25074927 Free PMC article.
-
Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence.Trends Microbiol. 2007 Feb;15(2):51-3. doi: 10.1016/j.tim.2006.12.005. Epub 2007 Jan 4. Trends Microbiol. 2007. PMID: 17207625 Free PMC article. Review.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
Cited by
-
Oligomeric State of β-Coronavirus Non-Structural Protein 10 Stimulators Studied by Small Angle X-ray Scattering.Int J Mol Sci. 2023 Sep 4;24(17):13649. doi: 10.3390/ijms241713649. Int J Mol Sci. 2023. PMID: 37686452 Free PMC article.
-
Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.Nucleic Acids Res. 2023 Jan 11;51(1):475-487. doi: 10.1093/nar/gkac1207. Nucleic Acids Res. 2023. PMID: 36546776 Free PMC article.
-
Comparative highlights on MERS-CoV, SARS-CoV-1, SARS-CoV-2, and NEO-CoV.EXCLI J. 2022 Sep 29;21:1245-1272. doi: 10.17179/excli2022-5355. eCollection 2022. EXCLI J. 2022. PMID: 36483910 Free PMC article. Review.
-
SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion.Viruses. 2022 Sep 8;14(9):1991. doi: 10.3390/v14091991. Viruses. 2022. PMID: 36146796 Free PMC article. Review.
-
A Temperature-Sensitive Recombinant of Avian Coronavirus Infectious Bronchitis Virus Provides Complete Protection against Homologous Challenge.J Virol. 2022 Sep 14;96(17):e0110022. doi: 10.1128/jvi.01100-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972294 Free PMC article.
References
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. - PubMed
-
- Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. - PMC - PubMed
-
- Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph Model 15:112-134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

