Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs
- PMID: 16873246
- PMCID: PMC1563791
- DOI: 10.1128/JVI.00467-06
Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs
Abstract
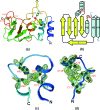
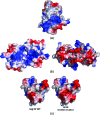
The severe acute respiratory syndrome coronavirus (SARS-CoV) possesses a large 29.7-kb positive-stranded RNA genome. The first open reading frame encodes replicase polyproteins 1a and 1ab, which are cleaved to generate 16 "nonstructural" proteins, nsp1 to nsp16, involved in viral replication and/or RNA processing. Among these, nsp10 plays a critical role in minus-strand RNA synthesis in a related coronavirus, murine hepatitis virus. Here, we report the crystal structure of SARS-CoV nsp10 at a resolution of 1.8 A as determined by single-wavelength anomalous dispersion using phases derived from hexatantalum dodecabromide. nsp10 is a single domain protein consisting of a pair of antiparallel N-terminal helices stacked against an irregular beta-sheet, a coil-rich C terminus, and two Zn fingers. nsp10 represents a novel fold and is the first structural representative of this family of Zn finger proteins found so far exclusively in coronaviruses. The first Zn finger coordinates a Zn2+ ion in a unique conformation. The second Zn finger, with four cysteines, is a distant member of the "gag-knuckle fold group" of Zn2+-binding domains and appears to maintain the structural integrity of the C-terminal tail. A distinct clustering of basic residues on the protein surface suggests a nucleic acid-binding function. Gel shift assays indicate that in isolation, nsp10 binds single- and double-stranded RNA and DNA with high-micromolar affinity and without obvious sequence specificity. It is possible that nsp10 functions within a larger RNA-binding protein complex. However, its exact role within the replicase complex is still not clear.
Figures


Similar articles
-
Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10.J Virol. 2006 Aug;80(16):7902-8. doi: 10.1128/JVI.00483-06. J Virol. 2006. PMID: 16873247 Free PMC article.
-
Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.PLoS Pathog. 2011 Oct;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. Epub 2011 Oct 13. PLoS Pathog. 2011. PMID: 22022266 Free PMC article.
-
Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):9436-41. doi: 10.1073/pnas.1508686112. Epub 2015 Jul 9. Proc Natl Acad Sci U S A. 2015. PMID: 26159422 Free PMC article.
-
The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing.Adv Virus Res. 2016;96:59-126. doi: 10.1016/bs.aivir.2016.08.008. Epub 2016 Sep 14. Adv Virus Res. 2016. PMID: 27712628 Free PMC article. Review.
-
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19.Cells. 2021 Apr 6;10(4):821. doi: 10.3390/cells10040821. Cells. 2021. PMID: 33917481 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion.Viruses. 2022 Sep 8;14(9):1991. doi: 10.3390/v14091991. Viruses. 2022. PMID: 36146796 Free PMC article. Review.
-
RNA-RNA and RNA-protein interactions in coronavirus replication and transcription.RNA Biol. 2011 Mar-Apr;8(2):237-48. doi: 10.4161/rna.8.2.14991. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21378501 Free PMC article. Review.
-
Coronavirus pathogenesis.Adv Virus Res. 2011;81:85-164. doi: 10.1016/B978-0-12-385885-6.00009-2. Adv Virus Res. 2011. PMID: 22094080 Free PMC article. Review.
-
Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes.J Biol Chem. 2014 Sep 12;289(37):25783-96. doi: 10.1074/jbc.M114.577353. Epub 2014 Jul 29. J Biol Chem. 2014. PMID: 25074927 Free PMC article.
-
Coronavirus infectious bronchitis virus non-structural proteins 8 and 12 form stable complex independent of the non-translated regions of viral RNA and other viral proteins.Virology. 2018 Jan 1;513:75-84. doi: 10.1016/j.virol.2017.10.004. Epub 2017 Oct 13. Virology. 2018. PMID: 29035788 Free PMC article.
References
-
- Banumathi, S., M. Dauter, and Z. Dauter. 2003. Phasing at high resolution using Ta6Br12 cluster. Acta Crystallogr. Sect. D. Biol. Crystallogr. 59:492-498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

