Gene expression analysis of zebrafish heart regeneration
- PMID: 16869712
- PMCID: PMC1523227
- DOI: 10.1371/journal.pbio.0040260
Gene expression analysis of zebrafish heart regeneration
Abstract
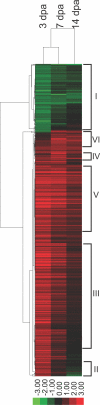
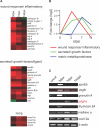
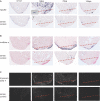
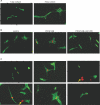
Mammalian hearts cannot regenerate. In contrast, zebrafish hearts regenerate even when up to 20% of the ventricle is amputated. The mechanism of zebrafish heart regeneration is not understood. To systematically characterize this process at the molecular level, we generated transcriptional profiles of zebrafish cardiac regeneration by microarray analyses. Distinct gene clusters were identified based on temporal expression patterns. Genes coding for wound response/inflammatory factors, secreted molecules, and matrix metalloproteinases are expressed in regenerating heart in sequential patterns. Comparisons of gene expression profiles between heart and fin regeneration revealed a set of regeneration core molecules as well as tissue-specific factors. The expression patterns of several secreted molecules around the wound suggest that they play important roles in heart regeneration. We found that both platelet-derived growth factor-a and -b (pdgf-a and pdgf-b) are upregulated in regenerating zebrafish hearts. PDGF-B homodimers induce DNA synthesis in adult zebrafish cardiomyocytes. In addition, we demonstrate that a chemical inhibitor of PDGF receptor decreases DNA synthesis of cardiomyocytes both in vitro and in vivo during regeneration. Our data indicate that zebrafish heart regeneration is associated with sequentially upregulated wound healing genes and growth factors and suggest that PDGF signaling is required.
Figures

Comment in
-
Regenerating zebrafish hearts reveal the molecular agents of repair.PLoS Biol. 2006 Aug;4(8):e281. doi: 10.1371/journal.pbio.0040281. Epub 2006 Aug 1. PLoS Biol. 2006. PMID: 20076625 Free PMC article. No abstract available.
Similar articles
-
Transcriptional profiling of caudal fin regeneration in zebrafish.ScientificWorldJournal. 2006 Jun 2;6 Suppl 1:38-54. doi: 10.1100/tsw.2006.326. ScientificWorldJournal. 2006. PMID: 17205186 Free PMC article.
-
Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration.PLoS One. 2013 Jun 26;8(6):e67266. doi: 10.1371/journal.pone.0067266. Print 2013. PLoS One. 2013. PMID: 23840646 Free PMC article.
-
Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration.Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13416-21. doi: 10.1073/pnas.1309810110. Epub 2013 Jul 30. Proc Natl Acad Sci U S A. 2013. PMID: 23901114 Free PMC article.
-
Cell migration during heart regeneration in zebrafish.Dev Dyn. 2016 Jul;245(7):774-87. doi: 10.1002/dvdy.24411. Epub 2016 May 10. Dev Dyn. 2016. PMID: 27085002 Free PMC article. Review.
-
Platelet-derived growth factor. Distinct signal transduction pathways associated with migration versus proliferation.Ann N Y Acad Sci. 1995 Sep 7;766:416-30. doi: 10.1111/j.1749-6632.1995.tb26691.x. Ann N Y Acad Sci. 1995. PMID: 7486687 Review.
Cited by
-
Ultrasound bio-microscopic image segmentation for evaluation of zebrafish cardiac function.IEEE Trans Ultrason Ferroelectr Freq Control. 2013 Apr;60(4):718-26. doi: 10.1109/TUFFC.2013.2620. IEEE Trans Ultrason Ferroelectr Freq Control. 2013. PMID: 23549532 Free PMC article.
-
Myocardial regeneration of the failing heart.Heart Fail Rev. 2013 Nov;18(6):815-33. doi: 10.1007/s10741-012-9348-5. Heart Fail Rev. 2013. PMID: 23001638 Review.
-
Zebrafish heart regeneration: Factors that stimulate cardiomyocyte proliferation.Semin Cell Dev Biol. 2020 Apr;100:3-10. doi: 10.1016/j.semcdb.2019.09.005. Epub 2019 Sep 25. Semin Cell Dev Biol. 2020. PMID: 31563389 Free PMC article. Review.
-
A histone demethylase is necessary for regeneration in zebrafish.Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19889-94. doi: 10.1073/pnas.0904132106. Epub 2009 Nov 6. Proc Natl Acad Sci U S A. 2009. PMID: 19897725 Free PMC article.
-
Platelet-derived growth factor receptor beta is critical for zebrafish intersegmental vessel formation.PLoS One. 2010 Jun 25;5(6):e11324. doi: 10.1371/journal.pone.0011324. PLoS One. 2010. PMID: 20593033 Free PMC article.
References
-
- Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. - PubMed
-
- Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. - PubMed
-
- Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. - PubMed
-
- Parmacek MS, Epstein JA. Pursuing cardiac progenitors: Regeneration redux. Cell. 2005;120:295–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

