Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase
- PMID: 16840321
- PMCID: PMC1563691
- DOI: 10.1128/JVI.02533-05
Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase
Abstract
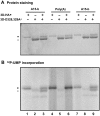
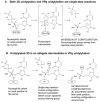
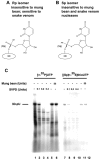
The 22-amino-acid protein VPg can be uridylylated in solution by purified poliovirus 3D polymerase in a template-dependent reaction thought to mimic primer formation during RNA amplification in infected cells. In the cell, the template used for the reaction is a hairpin RNA termed 2C-cre and, possibly, the poly(A) at the 3' end of the viral genome. Here, we identify several additional substrates for uridylylation by poliovirus 3D polymerase. In the presence of a 15-nucleotide (nt) RNA template, the poliovirus polymerase uridylylates other polymerase molecules in an intermolecular reaction that occurs in a single step, as judged by the chirality of the resulting phosphodiester linkage. Phosphate chirality experiments also showed that VPg uridylylation can occur by a single step; therefore, there is no obligatory uridylylated intermediate in the formation of uridylylated VPg. Other poliovirus proteins that could be uridylylated by 3D polymerase in solution were viral 3CD and 3AB proteins. Strong effects of both RNA and protein ligands on the efficiency and the specificity of the uridylylation reaction were observed: uridylylation of 3D polymerase and 3CD protein was stimulated by the addition of viral protein 3AB, and, when the template was poly(A) instead of the 15-nt RNA, the uridylylation of 3D polymerase itself became intramolecular instead of intermolecular. Finally, an antiuridine antibody identified uridylylated viral 3D polymerase and 3CD protein, as well as a 65- to 70-kDa host protein, in lysates of virus-infected human cells.
Figures



Similar articles
-
Allosteric effects of ligands and mutations on poliovirus RNA-dependent RNA polymerase.J Virol. 2005 Jun;79(12):7803-11. doi: 10.1128/JVI.79.12.7803-7811.2005. J Virol. 2005. PMID: 15919933 Free PMC article.
-
Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer.J Mol Biol. 2006 Mar 24;357(2):665-75. doi: 10.1016/j.jmb.2005.12.044. Epub 2006 Jan 5. J Mol Biol. 2006. PMID: 16427083
-
Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg.J Virol. 2000 Nov;74(22):10359-70. doi: 10.1128/jvi.74.22.10359-10370.2000. J Virol. 2000. PMID: 11044080 Free PMC article.
-
Uridylylation of the genome-linked protein of poliovirus in vitro is dependent upon an endogenous RNA template.Virus Res. 1987 Sep;8(3):193-204. doi: 10.1016/0168-1702(87)90015-3. Virus Res. 1987. PMID: 2825442 Review.
-
Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase.Curr Opin Struct Biol. 2009 Dec;19(6):752-8. doi: 10.1016/j.sbi.2009.10.016. Epub 2009 Nov 13. Curr Opin Struct Biol. 2009. PMID: 19914060 Review.
Cited by
-
Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein.Virology. 2008 Apr 25;374(1):33-49. doi: 10.1016/j.virol.2007.12.028. Epub 2008 Jan 30. Virology. 2008. PMID: 18234264 Free PMC article.
-
Coxsackievirus A6 Induces Cell Cycle Arrest in G0/G1 Phase for Viral Production.Front Cell Infect Microbiol. 2018 Aug 15;8:279. doi: 10.3389/fcimb.2018.00279. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30159255 Free PMC article.
-
Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants.J Virol. 2009 Sep;83(18):9370-87. doi: 10.1128/JVI.02076-08. Epub 2009 Jul 8. J Virol. 2009. PMID: 19587035 Free PMC article.
-
Novel roles of the picornaviral 3D polymerase in viral pathogenesis.Adv Virol. 2010 Jan 1;2010:368068. doi: 10.1155/2010/368068. Adv Virol. 2010. PMID: 20625447 Free PMC article.
-
Protein Nucleotidylylation in +ssRNA Viruses.Viruses. 2021 Aug 5;13(8):1549. doi: 10.3390/v13081549. Viruses. 2021. PMID: 34452414 Free PMC article. Review.
References
-
- Adler, S. P., D. Purich, and E. R. Stadtman. 1975. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl removing enzyme. J. Biol. Chem. 16:6264-6279. - PubMed
-
- Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′-end of poliovirus RNA. Cell 63:369-380. - PubMed
-
- Arabshahi, A., R. S. Brody, A. Smallwood, T.-C. Tsai, and P. A. Frey. 1986. Galactose-1-phosphate uridylyltransferase. Purification of the enzyme and stereochemical course of each step of the double-displacement mechanism. Biochemistry 25:5583-5589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

