Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors
- PMID: 16822840
- PMCID: PMC1593166
- DOI: 10.1091/mbc.e06-05-0470
Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors
Abstract
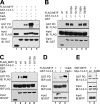
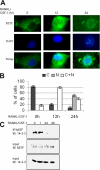
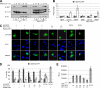
The microphthalmia-associated transcription factor (MITF) is required for terminal osteoclast differentiation and is a target for signaling pathways engaged by colony stimulating factor (CSF)-1 and receptor-activator of nuclear factor-kappaB ligand (RANKL). Work presented here demonstrates that MITF can shuttle from cytoplasm to nucleus dependent upon RANKL/CSF-1 action. 14-3-3 was identified as a binding partner of MITF in osteoclast precursors, and overexpression of 14-3-3 in a transgenic model resulted in increased cytosolic localization of MITF and decreased expression of MITF target genes. MITF/14-3-3 interaction was phosphorylation dependent, and Ser173 residue, within the minimal interaction region of amino acid residues 141-191, was required. The Cdc25C-associated kinase (C-TAK)1 interacted with an overlapping region of MITF. C-TAK1 increased MITF/14-3-3 complex formation and thus promoted cytoplasmic localization of MITF. C-TAK1 interaction was disrupted by RANKL/CSF-1 treatment. The results indicate that 14-3-3 regulates MITF activity by promoting the cytosolic localization of MITF in the absence of signals required for osteoclast differentiation. This work identifies a mechanism that regulates MITF activity in monocytic precursors that are capable of undergoing different terminal differentiation programs, and it provides a mechanism that allows committed precursors to rapidly respond to signals in the bone microenvironment to promote specifically osteoclast differentiation.
Figures


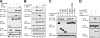
Similar articles
-
C-TAK1 interacts with microphthalmia-associated transcription factor, Mitf, but not the related family member Tfe3.Biochem Biophys Res Commun. 2010 Apr 16;394(4):890-5. doi: 10.1016/j.bbrc.2010.03.034. Epub 2010 Mar 7. Biochem Biophys Res Commun. 2010. PMID: 20214879 Free PMC article.
-
Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors.Mol Cell Biol. 2007 Jun;27(11):4018-27. doi: 10.1128/MCB.01839-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403896 Free PMC article.
-
Failure to Target RANKL Signaling Through p38-MAPK Results in Defective Osteoclastogenesis in the Microphthalmia Cloudy-Eyed Mutant.J Cell Physiol. 2016 Mar;231(3):630-40. doi: 10.1002/jcp.25108. J Cell Physiol. 2016. PMID: 26218069 Free PMC article.
-
The multifunctional protein fused in sarcoma (FUS) is a coactivator of microphthalmia-associated transcription factor (MITF).J Biol Chem. 2014 Jan 3;289(1):326-34. doi: 10.1074/jbc.M113.493874. Epub 2013 Nov 20. J Biol Chem. 2014. PMID: 24257758 Free PMC article.
-
Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function.Bone. 2004 Apr;34(4):689-96. doi: 10.1016/j.bone.2003.08.014. Bone. 2004. PMID: 15050900 Review.
Cited by
-
Overexpression of 14-3-3ζ Increases Brain Levels of C/EBP Homologous Protein CHOP.J Mol Neurosci. 2015 Jun;56(2):255-62. doi: 10.1007/s12031-015-0510-0. Epub 2015 Feb 18. J Mol Neurosci. 2015. PMID: 25854777
-
BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8668-E8677. doi: 10.1073/pnas.1810498115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150413 Free PMC article.
-
HDAC3 and HDAC7 have opposite effects on osteoclast differentiation.J Biol Chem. 2011 Apr 8;286(14):12056-65. doi: 10.1074/jbc.M110.216853. Epub 2011 Feb 15. J Biol Chem. 2011. PMID: 21324898 Free PMC article.
-
Enhancer variants reveal a conserved transcription factor network governed by PU.1 during osteoclast differentiation.Bone Res. 2018 Mar 28;6:8. doi: 10.1038/s41413-018-0011-1. eCollection 2018. Bone Res. 2018. PMID: 29619268 Free PMC article.
-
Versatility of 14-3-3 proteins and their roles in bone and joint-related diseases.Bone Res. 2024 Oct 15;12(1):58. doi: 10.1038/s41413-024-00370-4. Bone Res. 2024. PMID: 39406741 Free PMC article. Review.
References
-
- Aitken A., Howell S., Jones D., Madrazo J., Martin H., Patel Y., Robinson K. Post-translationally modified 14-3-3 isoforms and inhibition of PKC. Mol. Cell Biochem. 1995;149–150:41–49. - PubMed
-
- Andrin C., Hendzel M. J. F-actin-dependent insolubility of chromatin-modifying components. J. Biol. Chem. 2004;279:25017–25023. - PubMed
-
- Bachmann M., Hennemann H., Xing P. X., Hoffmann I., Moroy T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J. Biol. Chem. 2004;279:48319–48328. - PubMed
-
- Boyle W. J., Simonet W. S., Lacey D. L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. - PubMed
-
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

