Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer
- PMID: 16798872
- PMCID: PMC1502457
- DOI: 10.1073/pnas.0604232103
Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer
Abstract
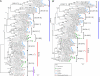
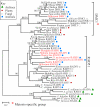
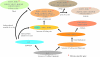
The bacterial recA gene and its eukaryotic homolog RAD51 are important for DNA repair, homologous recombination, and genome stability. Members of the recA/RAD51 family have functions that have differentiated during evolution. However, the evolutionary history and relationships of these members remains unclear. Homolog searches in prokaryotes and eukaryotes indicated that most eubacteria contain only one recA. However, many archaeal species have two recA/RAD51 homologs (RADA and RADB), and eukaryotes possess multiple members (RAD51, RAD51B, RAD51C, RAD51D, DMC1, XRCC2, XRCC3, and recA). Phylogenetic analyses indicated that the recA/RAD51 family can be divided into three subfamilies: (i) RADalpha, with highly conserved functions; (ii) RADbeta, with relatively divergent functions; and (iii) recA, functioning in eubacteria and eukaryotic organelles. The RADalpha and RADbeta subfamilies each contain archaeal and eukaryotic members, suggesting that a gene duplication occurred before the archaea/eukaryote split. In the RADalpha subfamily, eukaryotic RAD51 and DMC1 genes formed two separate monophyletic groups when archaeal RADA genes were used as an outgroup. This result suggests that another duplication event occurred in the early stage of eukaryotic evolution, producing the DMC1 clade with meiosis-specific genes. The RADbeta subfamily has a basal archaeal clade and five eukaryotic clades, suggesting that four eukaryotic duplication events occurred before animals and plants diverged. The eukaryotic recA genes were detected in plants and protists and showed strikingly high levels of sequence similarity to recA genes from proteobacteria or cyanobacteria. These results suggest that endosymbiotic transfer of recA genes occurred from mitochondria and chloroplasts to nuclear genomes of ancestral eukaryotes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


Similar articles
-
Reevaluation of the evolutionary events within recA/RAD51 phylogeny.BMC Genomics. 2013 Apr 10;14:240. doi: 10.1186/1471-2164-14-240. BMC Genomics. 2013. PMID: 23574621 Free PMC article.
-
Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum, and phylogenetic analysis of eukaryotic recA homologs.Curr Genet. 1997 Feb;31(2):144-57. doi: 10.1007/s002940050189. Curr Genet. 1997. PMID: 9021132
-
Network Analysis of RAD51 Proteins in Metazoa and the Evolutionary Relationships With Their Archaeal Homologs.Front Genet. 2018 Sep 26;9:383. doi: 10.3389/fgene.2018.00383. eCollection 2018. Front Genet. 2018. PMID: 30319685 Free PMC article.
-
Role of recA/RAD51 family proteins in mammals.Acta Med Okayama. 2005 Feb;59(1):1-9. doi: 10.18926/AMO/31987. Acta Med Okayama. 2005. PMID: 15902993 Review.
-
Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination.Cytogenet Genome Res. 2004;107(3-4):201-7. doi: 10.1159/000080598. Cytogenet Genome Res. 2004. PMID: 15467365 Review.
Cited by
-
Expanded roles for the MutL family of DNA mismatch repair proteins.Yeast. 2021 Jan;38(1):39-53. doi: 10.1002/yea.3512. Epub 2020 Jul 30. Yeast. 2021. PMID: 32652606 Free PMC article. Review.
-
Application of RNAi-induced gene expression profiles for prognostic prediction in breast cancer.Genome Med. 2016 Oct 27;8(1):114. doi: 10.1186/s13073-016-0363-3. Genome Med. 2016. PMID: 27788678 Free PMC article.
-
RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome.Plant Physiol. 2012 May;159(1):211-26. doi: 10.1104/pp.112.194720. Epub 2012 Mar 13. Plant Physiol. 2012. PMID: 22415515 Free PMC article.
-
Ubiquitylation of Rad51d Mediated by E3 Ligase Rnf138 Promotes the Homologous Recombination Repair Pathway.PLoS One. 2016 May 19;11(5):e0155476. doi: 10.1371/journal.pone.0155476. eCollection 2016. PLoS One. 2016. PMID: 27195665 Free PMC article.
-
The RAD51 and DMC1 homoeologous genes of bread wheat: cloning, molecular characterization and expression analysis.BMC Res Notes. 2010 Sep 29;3:245. doi: 10.1186/1756-0500-3-245. BMC Res Notes. 2010. PMID: 20920212 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

