microRNAs exhibit high frequency genomic alterations in human cancer
- PMID: 16754881
- PMCID: PMC1474008
- DOI: 10.1073/pnas.0508889103
microRNAs exhibit high frequency genomic alterations in human cancer
Abstract
MicroRNAs (miRNAs) are endogenous noncoding RNAs, which negatively regulate gene expression. To determine genomewide miRNA DNA copy number abnormalities in cancer, 283 known human miRNA genes were analyzed by high-resolution array-based comparative genomic hybridization in 227 human ovarian cancer, breast cancer, and melanoma specimens. A high proportion of genomic loci containing miRNA genes exhibited DNA copy number alterations in ovarian cancer (37.1%), breast cancer (72.8%), and melanoma (85.9%), where copy number alterations observed in >15% tumors were considered significant for each miRNA gene. We identified 41 miRNA genes with gene copy number changes that were shared among the three cancer types (26 with gains and 15 with losses) as well as miRNA genes with copy number changes that were unique to each tumor type. Importantly, we show that miRNA copy changes correlate with miRNA expression. Finally, we identified high frequency copy number abnormalities of Dicer1, Argonaute2, and other miRNA-associated genes in breast and ovarian cancer as well as melanoma. These findings support the notion that copy number alterations of miRNAs and their regulatory genes are highly prevalent in cancer and may account partly for the frequent miRNA gene deregulation reported in several tumor types.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
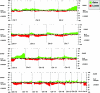


Similar articles
-
Are microRNAs located in genomic regions associated with cancer?Br J Cancer. 2006 Nov 20;95(10):1415-8. doi: 10.1038/sj.bjc.6603381. Epub 2006 Sep 26. Br J Cancer. 2006. PMID: 17003783 Free PMC article.
-
High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization.Breast Cancer Res. 2005;7(6):R1186-98. doi: 10.1186/bcr1356. Epub 2005 Nov 24. Breast Cancer Res. 2005. PMID: 16457699 Free PMC article.
-
Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma.Cancer Genet. 2011 Mar;204(3):138-46. doi: 10.1016/j.cancergen.2010.12.012. Cancer Genet. 2011. PMID: 21504713
-
[Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese.
-
Array comparative genomic hybridization and its applications in cancer.Nat Genet. 2005 Jun;37 Suppl:S11-7. doi: 10.1038/ng1569. Nat Genet. 2005. PMID: 15920524 Review.
Cited by
-
The miR-200 Family: Versatile Players in Epithelial Ovarian Cancer.Int J Mol Sci. 2015 Jul 24;16(8):16833-47. doi: 10.3390/ijms160816833. Int J Mol Sci. 2015. PMID: 26213923 Free PMC article. Review.
-
miRNAs in human cancer.Methods Mol Biol. 2012;822:295-306. doi: 10.1007/978-1-61779-427-8_21. Methods Mol Biol. 2012. PMID: 22144208 Free PMC article.
-
Genome-wide analysis of miRNA signature differentially expressed in doxorubicin-resistant and parental human hepatocellular carcinoma cell lines.PLoS One. 2013;8(1):e54111. doi: 10.1371/journal.pone.0054111. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23359607 Free PMC article.
-
The Association between Five Genetic Variants in MicroRNAs (rs2910164, rs11614913, rs3746444, rs11134527, and rs531564) and Cervical Cancer Risk: A Meta-Analysis.Biomed Res Int. 2021 Mar 15;2021:9180874. doi: 10.1155/2021/9180874. eCollection 2021. Biomed Res Int. 2021. PMID: 33816633 Free PMC article.
-
An integrated analysis of miRNA and gene copy numbers in xenografts of Ewing's sarcoma.J Exp Clin Cancer Res. 2012 Mar 20;31(1):24. doi: 10.1186/1756-9966-31-24. J Exp Clin Cancer Res. 2012. PMID: 22429812 Free PMC article.
References
-
- Lee R. C., Feinbaum R. L., Ambros V. Cell. 1993;75:843–854. - PubMed
-
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Science. 2001;294:853–858. - PubMed
-
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. Science. 2001;294:858–862. - PubMed
-
- Lee R. C., Ambros V. Science. 2001;294:862–864. - PubMed
-
- Bartel D. P. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

