STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis
- PMID: 16751345
- PMCID: PMC1488911
- DOI: 10.1105/tpc.106.042184
STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis
Abstract
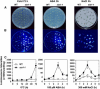
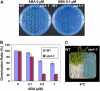
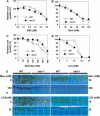
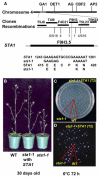
In plants, many gene transcripts are very unstable, which is important for the tight control of their temporal and spatial expression patterns. To identify cellular factors controlling the stability of unstable mRNAs in plants, we used luciferase imaging in Arabidopsis thaliana to isolate a recessive mutant, stabilized1-1 (sta1-1), with enhanced stability of the normally unstable luciferase transcript. The sta1-1 mutation also causes the stabilization of some endogenous gene transcripts and has a range of developmental and stress response phenotypes. STA1 encodes a nuclear protein similar to the human U5 small ribonucleoprotein-associated 102-kD protein and to the yeast pre-mRNA splicing factors Prp1p and Prp6p. STA1 expression is upregulated by cold stress, and the sta1-1 mutant is defective in the splicing of the cold-induced COR15A gene. Our results show that STA1 is a pre-mRNA splicing factor required not only for splicing but also for the turnover of unstable transcripts and that it has an important role in plant responses to abiotic stresses.
Figures


Similar articles
-
STABILIZED1 Modulates Pre-mRNA Splicing for Thermotolerance.Plant Physiol. 2017 Apr;173(4):2370-2382. doi: 10.1104/pp.16.01928. Epub 2017 Feb 21. Plant Physiol. 2017. PMID: 28223317 Free PMC article.
-
Genetic Screening for Arabidopsis Mutants Defective in STA1 Regulation under Thermal Stress Implicates the Existence of Regulators of Its Specific Expression, and the Genetic Interactions in the Stress Signaling Pathways.Front Plant Sci. 2016 May 10;7:618. doi: 10.3389/fpls.2016.00618. eCollection 2016. Front Plant Sci. 2016. PMID: 27242824 Free PMC article.
-
STABILIZED1 as a heat stress-specific splicing factor in Arabidopsis thaliana.Plant Signal Behav. 2018 Feb 1;13(2):e1432955. doi: 10.1080/15592324.2018.1432955. Epub 2018 Feb 16. Plant Signal Behav. 2018. PMID: 29381447 Free PMC article.
-
Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):875-89. doi: 10.1002/wrna.98. Epub 2011 Jul 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21766458 Review.
-
Role of plant RNA-binding proteins in development, stress response and genome organization.Trends Plant Sci. 2009 Apr;14(4):229-36. doi: 10.1016/j.tplants.2009.01.007. Epub 2009 Mar 13. Trends Plant Sci. 2009. PMID: 19285908 Review.
Cited by
-
Characterization of barley Prp1 gene and its expression during seed development and under abiotic stress.Genetica. 2011 Oct;139(10):1283-92. doi: 10.1007/s10709-012-9630-4. Genetica. 2011. PMID: 22290495
-
A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis.Plant Cell. 2013 Jan;25(1):342-56. doi: 10.1105/tpc.112.108340. Epub 2013 Jan 31. Plant Cell. 2013. PMID: 23371945 Free PMC article.
-
Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3.BMC Genomics. 2010 Jan 28;11:69. doi: 10.1186/1471-2164-11-69. BMC Genomics. 2010. PMID: 20105335 Free PMC article.
-
Breeding approaches and genomics technologies to increase crop yield under low-temperature stress.Plant Cell Rep. 2017 Jan;36(1):1-35. doi: 10.1007/s00299-016-2073-0. Epub 2016 Nov 22. Plant Cell Rep. 2017. PMID: 27878342 Review.
-
Alternative splicing of transcription factors in plant responses to low temperature stress: mechanisms and functions.Planta. 2013 Jun;237(6):1415-24. doi: 10.1007/s00425-013-1882-4. Epub 2013 Apr 28. Planta. 2013. PMID: 23624977 Free PMC article. Review.
References
-
- Abler, M.L., and Green, P.J. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32 63–78. - PubMed
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. - PubMed
-
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

