Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2
- PMID: 16738331
- PMCID: PMC1489137
- DOI: 10.1128/MCB.01830-05
Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2
Abstract
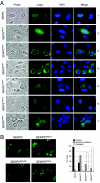
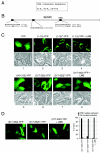
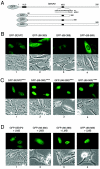
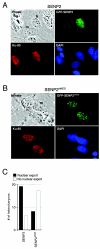
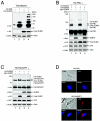
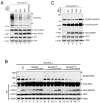
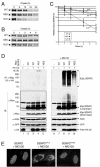
Small ubiquitin-related modifier (SUMO) proteins are conjugated to numerous polypeptides in cells, and attachment of SUMO plays important roles in regulating the activity, stability, and subcellular localization of modified proteins. SUMO modification of proteins is a dynamic and reversible process. A family of SUMO-specific proteases catalyzes the deconjugation of SUMO-modified proteins. Members of the Sentrin (also known as SUMO)-specific protease (SENP) family have been characterized with unique subcellular localizations. However, little is known about the functional significance of or the regulatory mechanism derived from the specific localizations of the SENPs. Here we identify a bipartite nuclear localization signal (NLS) and a CRM1-dependent nuclear export signal (NES) in the SUMO protease SENP2. Both the NLS and the NES are located in the nonhomologous domains of SENP2 and are not conserved among other members of the SENP family. Using a series of SENP2 mutants and a heterokaryon assay, we demonstrate that SENP2 shuttles between the nucleus and the cytoplasm and that the shuttling is blocked by mutations in the NES or by treating cells with leptomycin B. We show that SENP2 can be polyubiquitinated in vivo and degraded through proteolysis. Restricting SENP2 in the nucleus by mutations in the NES impairs its polyubiquitination, whereas a cytoplasm-localized SENP2 made by introducing mutations in the NLS can be efficiently polyubiquitinated, suggesting that SENP2 is ubiquitinated in the cytoplasm. Finally, treating cells with MG132 leads to accumulation of polyubiquitinated SENP2, indicating that SENP2 is degraded through the 26S proteolysis pathway. Thus, the function of SENP2 is regulated by both nucleocytoplasmic shuttling and polyubiquitin-mediated degradation.
Figures

Similar articles
-
Association of the human SUMO-1 protease SENP2 with the nuclear pore.J Biol Chem. 2002 May 31;277(22):19961-6. doi: 10.1074/jbc.M201799200. Epub 2002 Mar 14. J Biol Chem. 2002. PMID: 11896061
-
Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination.Mol Cell Biol. 2003 Sep;23(18):6396-405. doi: 10.1128/MCB.23.18.6396-6405.2003. Mol Cell Biol. 2003. PMID: 12944468 Free PMC article.
-
SUMO1/sentrin/SMT3 specific peptidase 2 modulates target molecules and its corresponding functions.Biochimie. 2018 Sep;152:6-13. doi: 10.1016/j.biochi.2018.06.007. Epub 2018 Jun 21. Biochimie. 2018. PMID: 29908207 Review.
-
A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex.Structure. 2004 Aug;12(8):1519-31. doi: 10.1016/j.str.2004.05.023. Structure. 2004. PMID: 15296745
-
SUMO-specific proteases and isopeptidases of the SENP family at a glance.J Cell Sci. 2018 Mar 20;131(6):jcs211904. doi: 10.1242/jcs.211904. J Cell Sci. 2018. PMID: 29559551 Review.
Cited by
-
Regulation of organic anion transporters: Role in physiology, pathophysiology, and drug elimination.Pharmacol Ther. 2021 Jan;217:107647. doi: 10.1016/j.pharmthera.2020.107647. Epub 2020 Aug 3. Pharmacol Ther. 2021. PMID: 32758646 Free PMC article. Review.
-
Distribution and paralogue specificity of mammalian deSUMOylating enzymes.Biochem J. 2010 Sep 1;430(2):335-44. doi: 10.1042/BJ20100504. Biochem J. 2010. PMID: 20590526 Free PMC article.
-
The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition.Nat Rev Mol Cell Biol. 2010 Dec;11(12):861-71. doi: 10.1038/nrm3011. Nat Rev Mol Cell Biol. 2010. PMID: 21102611 Free PMC article. Review.
-
Starting and stopping SUMOylation. What regulates the regulator?Chromosoma. 2013 Dec;122(6):451-63. doi: 10.1007/s00412-013-0422-0. Epub 2013 Jun 28. Chromosoma. 2013. PMID: 23812602 Review.
-
SENP1 and SENP2 regulate SUMOylation of amyloid precursor protein.Heliyon. 2018 Apr 13;4(4):e00601. doi: 10.1016/j.heliyon.2018.e00601. eCollection 2018 Apr. Heliyon. 2018. PMID: 29862363 Free PMC article.
References
-
- Alarcon-Vargas, D., and Z. Ronai. 2002. SUMO in cancer—wrestlers wanted. Cancer Biol. Ther. 1:237-242. - PubMed
-
- Bailey, D., and P. O'Hare. 2004. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279:692-703. - PubMed
-
- Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. - PubMed
-
- Bohren, K. M., V. Nadkarni, J. H. Song, K. H. Gabbay, and D. Owerbach. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279:27233-27238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
