The dsRNA binding site of human Toll-like receptor 3
- PMID: 16720699
- PMCID: PMC1482657
- DOI: 10.1073/pnas.0603245103
The dsRNA binding site of human Toll-like receptor 3
Abstract
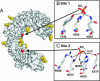
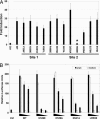
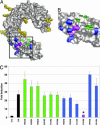
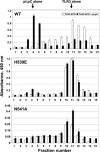
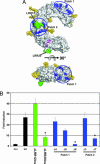
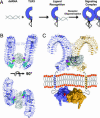
Pathogen recognition by Toll-like receptors (TLRs) initiates innate immune responses that are essential for inhibiting pathogen dissemination and for the development of acquired immunity. The TLRs recognize pathogens with their N-terminal ectodomains (ECD), but the molecular basis for this recognition is not known. Recently we reported the x-ray structure for unliganded TLR3-ECD; however, it has proven difficult to obtain a crystal structure of TLR3 with its ligand, dsRNA. We have now located the TLR3 ligand binding site by mutational analysis. More than 50 single-residue mutations have been generated throughout the TLR3-ECD, but only two, H539E and N541A, resulted in the loss of TLR3 activation and ligand binding functions. These mutations locate the dsRNA binding site on the glycan-free, lateral surface of TLR3 toward the C terminus and suggest a model for dsRNA binding and TLR3 activation.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Similar articles
-
N-terminal binding site in the human toll-like receptor 3 ectodomain.Nucleic Acids Symp Ser (Oxf). 2007;(51):405-6. doi: 10.1093/nass/nrm203. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029758
-
Structural basis of toll-like receptor 3 signaling with double-stranded RNA.Science. 2008 Apr 18;320(5874):379-81. doi: 10.1126/science.1155406. Science. 2008. PMID: 18420935 Free PMC article.
-
Significance of the N-terminal histidine-rich region for the function of the human toll-like receptor 3 ectodomain.Nucleic Acids Symp Ser (Oxf). 2008;(52):203-4. doi: 10.1093/nass/nrn103. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776324
-
The toll-like receptor 3:dsRNA signaling complex.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):667-74. doi: 10.1016/j.bbagrm.2009.06.005. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19595807 Free PMC article. Review.
-
Beyond dsRNA: Toll-like receptor 3 signalling in RNA-induced immune responses.Biochem J. 2014 Mar 1;458(2):195-201. doi: 10.1042/BJ20131492. Biochem J. 2014. PMID: 24524192 Review.
Cited by
-
TLR3 immunity to infection in mice and humans.Curr Opin Immunol. 2013 Feb;25(1):19-33. doi: 10.1016/j.coi.2012.11.001. Epub 2013 Jan 3. Curr Opin Immunol. 2013. PMID: 23290562 Free PMC article. Review.
-
Pattern recognition receptors require N-glycosylation to mediate plant immunity.J Biol Chem. 2010 Feb 12;285(7):4629-36. doi: 10.1074/jbc.M109.063073. Epub 2009 Dec 11. J Biol Chem. 2010. PMID: 20007973 Free PMC article.
-
Immunomodulatory effects of dsRNA and its potential as vaccine adjuvant.J Biomed Biotechnol. 2010;2010:690438. doi: 10.1155/2010/690438. Epub 2010 Jul 5. J Biomed Biotechnol. 2010. PMID: 20671921 Free PMC article. Review.
-
Effects of bacterial and viral pathogen-associated molecular patterns (PAMPs) on multidrug resistance (MDR) transporters in brain endothelial cells of the developing human blood-brain barrier.Fluids Barriers CNS. 2023 Jan 31;20(1):8. doi: 10.1186/s12987-023-00409-4. Fluids Barriers CNS. 2023. PMID: 36721242 Free PMC article.
-
Trafficking of endosomal Toll-like receptors.Trends Cell Biol. 2014 Jun;24(6):360-9. doi: 10.1016/j.tcb.2013.12.002. Epub 2014 Jan 15. Trends Cell Biol. 2014. PMID: 24439965 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

