Involvement of the SMRT/NCoR-HDAC3 complex in transcriptional repression by the CNOT2 subunit of the human Ccr4-Not complex
- PMID: 16712523
- PMCID: PMC1559471
- DOI: 10.1042/BJ20060406
Involvement of the SMRT/NCoR-HDAC3 complex in transcriptional repression by the CNOT2 subunit of the human Ccr4-Not complex
Abstract
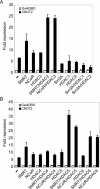
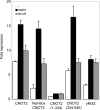
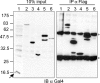
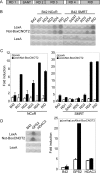
In eukaryotic cells, the Ccr4-Not complex can regulate mRNA metabolism at various levels. Previously, we showed that promoter targeting of the CNOT2 subunit resulted in strong repression of RNA polymerase II transcription, which was sensitive to the HDAC (histone deacetylase) inhibitor, trichostatin A [Zwartjes, Jayne, van den Berg and Timmers (2004) J. Biol. Chem. 279, 10848-10854]. In the present study, the cofactor requirement for CNOT2-mediated repression was investigated. We found that coexpression of SMRT (silencing mediator for retinoic acid receptor and thyroid-hormone receptor) or NCoR (nuclear hormone receptor co-repressor) in combination with HDAC3 (or HDAC5 and HDAC6) augmented the repression by CNOT2. This repressive effect is mediated by the conserved Not-Box, which resides at the C-terminus of CNOT2 proteins. We observed physical interactions of CNOT2 with several subunits of the SMRT/NCoR-HDAC3 complex. Our results show that the SMRT/NCoR-HDAC3 complex is a cofactor of CNOT2-mediated repression and suggest that transcriptional regulation by the Ccr4-Not complex involves regulation of chromatin modification.
Figures
Similar articles
-
Bag-1M inhibits the transactivation of the glucocorticoid receptor via recruitment of corepressors.FEBS Lett. 2009 Aug 6;583(15):2451-6. doi: 10.1016/j.febslet.2009.07.010. Epub 2009 Jul 15. FEBS Lett. 2009. PMID: 19595997
-
The functional relationship between co-repressor N-CoR and SMRT in mediating transcriptional repression by thyroid hormone receptor alpha.Biochem J. 2008 Apr 1;411(1):19-26. doi: 10.1042/BJ20071393. Biochem J. 2008. PMID: 18052923
-
Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway.EMBO J. 2000 Aug 1;19(15):4074-90. doi: 10.1093/emboj/19.15.4074. EMBO J. 2000. PMID: 10921888 Free PMC article.
-
Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
-
Coactivator and corepressor complexes in nuclear receptor function.Curr Opin Genet Dev. 1999 Apr;9(2):140-7. doi: 10.1016/S0959-437X(99)80021-5. Curr Opin Genet Dev. 1999. PMID: 10322133 Review.
Cited by
-
Ccr4-Not complex: the control freak of eukaryotic cells.Crit Rev Biochem Mol Biol. 2012 Jul-Aug;47(4):315-33. doi: 10.3109/10409238.2012.667214. Epub 2012 Mar 15. Crit Rev Biochem Mol Biol. 2012. PMID: 22416820 Free PMC article. Review.
-
Single-cell multi-omics profiling reveals key regulatory mechanisms that poise germinal vesicle oocytes for maturation in pigs.Cell Mol Life Sci. 2023 Jul 22;80(8):222. doi: 10.1007/s00018-023-04873-x. Cell Mol Life Sci. 2023. PMID: 37480402 Free PMC article.
-
Quantitative proteomic characterization and comparison of T helper 17 and induced regulatory T cells.PLoS Biol. 2018 May 31;16(5):e2004194. doi: 10.1371/journal.pbio.2004194. eCollection 2018 May. PLoS Biol. 2018. PMID: 29851958 Free PMC article.
-
Novel and Rare Fusion Transcripts Involving Transcription Factors and Tumor Suppressor Genes in Acute Myeloid Leukemia.Cancers (Basel). 2019 Dec 5;11(12):1951. doi: 10.3390/cancers11121951. Cancers (Basel). 2019. PMID: 31817495 Free PMC article.
-
Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa.Plant Mol Biol. 2014 Jul;85(4-5):443-58. doi: 10.1007/s11103-014-0196-7. Epub 2014 May 8. Plant Mol Biol. 2014. PMID: 24805883
References
-
- Martens J. A., Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. - PubMed
-
- Berger S. L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. - PubMed
-
- Collart M. A., Timmers H. T. The eukaryotic Ccr4–not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. - PubMed
-
- Denis C. L., Chen J. The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. - PubMed
-
- Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

