The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins
- PMID: 16702539
- PMCID: PMC1472451
- DOI: 10.1073/pnas.0602902103
The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins
Abstract
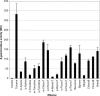
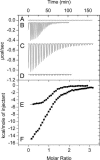
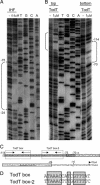
The TodS and TodT proteins form a previously unrecognized and highly specific two-component regulatory system in which the TodS sensor protein contains two input domains, each of which are coupled to a histidine kinase domain. This system regulates the expression of the genes involved in the degradation of toluene, benzene, and ethylbenzene through the toluene dioxygenase pathway. In contrast to the narrow substrate range of this catabolic pathway, the TodS effector profile is broad. TodS has basal autophosphorylation activity in vitro, which is enhanced by the presence of effectors. Toluene binds to TodS with high affinity (Kd = 684 +/- 13 nM) and 1:1 stoichiometry. The analysis of the truncated variants of TodS reveals that toluene binds to the N-terminal input domain (Kd = 2.3 +/- 0.1 microM) but not to the C-terminal half. TodS transphosphorylates TodT, which binds to two highly similar DNA binding sites at base pairs -107 and -85 of the promoter. Integration host factor (IHF) plays a crucial role in the activation process and binds between the upstream TodT boxes and the -10 hexamer region. In an IHF-deficient background, expression from the tod promoter drops 8-fold. In vitro transcription assays confirmed the role determined in vivo for TodS, TodT, and IHF. A functional model is presented in which IHF favors the contact between the TodT activator, bound further upstream, and the alpha-subunit of RNA polymerase bound to the downstream promoter element. Once these contacts are established, the tod operon is efficiently transcribed.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Hierarchical binding of the TodT response regulator to its multiple recognition sites at the tod pathway operon promoter.J Mol Biol. 2008 Feb 15;376(2):325-37. doi: 10.1016/j.jmb.2007.12.004. Epub 2007 Dec 8. J Mol Biol. 2008. PMID: 18166197
-
Two levels of cooperativeness in the binding of TodT to the tod operon promoter.J Mol Biol. 2008 Dec 31;384(5):1037-47. doi: 10.1016/j.jmb.2008.10.011. Epub 2008 Oct 11. J Mol Biol. 2008. PMID: 18950641
-
Construction of a prototype two-component system from the phosphorelay system TodS/TodT.Protein Eng Des Sel. 2012 Apr;25(4):159-69. doi: 10.1093/protein/gzs001. Epub 2012 Feb 3. Protein Eng Des Sel. 2012. PMID: 22308529
-
Catabolite repression of the TodS/TodT two-component system and effector-dependent transphosphorylation of TodT as the basis for toluene dioxygenase catabolic pathway control.J Bacteriol. 2010 Aug;192(16):4246-50. doi: 10.1128/JB.00379-10. Epub 2010 Jun 11. J Bacteriol. 2010. PMID: 20543072 Free PMC article.
-
Look, no hands! Unconventional transcriptional activators in bacteria.Trends Microbiol. 2007 Dec;15(12):530-7. doi: 10.1016/j.tim.2007.09.008. Epub 2007 Nov 9. Trends Microbiol. 2007. PMID: 17997097 Review.
Cited by
-
Fluoro-recognition: New in vivo fluorescent assay for toluene dioxygenase probing induction by and metabolism of polyfluorinated compounds.Environ Microbiol. 2022 Nov;24(11):5202-5216. doi: 10.1111/1462-2920.16187. Epub 2022 Oct 17. Environ Microbiol. 2022. PMID: 36054238 Free PMC article.
-
Construction and application of an Escherichia coli bioreporter for aniline and chloroaniline detection.J Ind Microbiol Biotechnol. 2012 Dec;39(12):1801-10. doi: 10.1007/s10295-012-1180-3. Epub 2012 Aug 15. J Ind Microbiol Biotechnol. 2012. PMID: 22892886
-
The pH Robustness of Bacterial Sensing.mBio. 2022 Oct 26;13(5):e0165022. doi: 10.1128/mbio.01650-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154178 Free PMC article.
-
Microcalorimetry: a response to challenges in modern biotechnology.Microb Biotechnol. 2008 Mar;1(2):126-36. doi: 10.1111/j.1751-7915.2007.00013.x. Microb Biotechnol. 2008. PMID: 21261830 Free PMC article. Review.
-
Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis.J Bacteriol. 2008 Nov;190(21):7147-56. doi: 10.1128/JB.00952-08. Epub 2008 Sep 5. J Bacteriol. 2008. PMID: 18776017 Free PMC article.
References
-
- Zylstra G. J., Gibson D. T. J. Biol. Chem. 1989;264:14940–14941. - PubMed
-
- Mosqueda G., Ramos-González M. I., Ramos J. L. Gene. 1999;232:69–76. - PubMed
-
- Choi E. N., Cho M. C., Kim Y., Kim C.-K., Lee K. Microbiology. 2003;149:795–805. - PubMed
-
- Domínguez-Cuevas & Marqués S. Pseudomonas. Vol. 2. Kluwer, Dordrecht: The Netherlands; 2004. pp. 319–345.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

