Evidence for functional protein interactions required for poliovirus RNA replication
- PMID: 16699013
- PMCID: PMC1472133
- DOI: 10.1128/JVI.02684-05
Evidence for functional protein interactions required for poliovirus RNA replication
Abstract
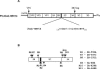
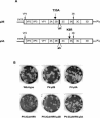
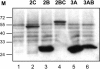
Poliovirus protein 2C contains a predicted N-terminal amphipathic helix that mediates association of the protein with the membranes of the viral RNA replication complex. A chimeric virus that contains sequences encoding the 18-residue core from the orthologous amphipathic helix from human rhinovirus type 14 (HRV14) was constructed. The chimeric virus exhibited defects in viral RNA replication and produced minute plaques on HeLa cell monolayers. Large plaque variants that contained mutations within the 2C-encoding region were generated upon subsequent passage. However, the majority of viruses that emerged with improved growth properties contained no changes in the region encoding 2C. Sequence analysis and reconstruction of genomes with individual mutations revealed changes in 3A or 2B sequences that compensated for the HRV14 amphipathic helix in the polio 2C-containing proteins, implying functional interactions among these proteins during the replication process. Direct binding between these viral proteins was confirmed by mammalian cell two-hybrid analysis.
Figures
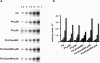

Similar articles
-
Testing the modularity of the N-terminal amphipathic helix conserved in picornavirus 2C proteins and hepatitis C NS5A protein.Virology. 2006 Jan 20;344(2):453-67. doi: 10.1016/j.virol.2005.08.044. Epub 2005 Oct 14. Virology. 2006. PMID: 16226781 Free PMC article.
-
An Amphipathic Alpha-Helix Domain from Poliovirus 2C Protein Tubulate Lipid Vesicles.Viruses. 2020 Dec 18;12(12):1466. doi: 10.3390/v12121466. Viruses. 2020. PMID: 33353144 Free PMC article.
-
Chimeric coxsackie B3 virus genomes that express hybrid coxsackievirus-poliovirus 2B proteins: functional dissection of structural domains involved in RNA replication.J Gen Virol. 1997 Aug;78 ( Pt 8):1833-40. doi: 10.1099/0022-1317-78-8-1833. J Gen Virol. 1997. PMID: 9266977
-
Amino terminal regions of poliovirus 2C protein mediate membrane binding.Virology. 1995 Apr 20;208(2):540-53. doi: 10.1006/viro.1995.1185. Virology. 1995. PMID: 7747426
-
IRES-controlled protein synthesis and genome replication of poliovirus.Arch Virol Suppl. 1994;9:279-89. doi: 10.1007/978-3-7091-9326-6_28. Arch Virol Suppl. 1994. PMID: 8032259 Review.
Cited by
-
Alanine scanning of poliovirus 2CATPase reveals new genetic evidence that capsid protein/2CATPase interactions are essential for morphogenesis.J Virol. 2012 Sep;86(18):9964-75. doi: 10.1128/JVI.00914-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761387 Free PMC article.
-
Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells.J Virol. 2011 May;85(9):4284-96. doi: 10.1128/JVI.02398-10. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345960 Free PMC article.
-
Polyprotein context regulates the activity of poliovirus 2CATPase bound to bilayer nanodiscs.J Virol. 2013 May;87(10):5994-6004. doi: 10.1128/JVI.03491-12. Epub 2013 Mar 20. J Virol. 2013. PMID: 23514879 Free PMC article.
-
The cis-acting replication elements define human enterovirus and rhinovirus species.RNA. 2008 Aug;14(8):1568-78. doi: 10.1261/rna.1031408. Epub 2008 Jun 9. RNA. 2008. PMID: 18541697 Free PMC article.
-
Investigating the role of viral integral membrane proteins in promoting the assembly of nepovirus and comovirus replication factories.Front Plant Sci. 2013 Jan 29;3:313. doi: 10.3389/fpls.2012.00313. eCollection 2012. Front Plant Sci. 2013. PMID: 23439982 Free PMC article.
References
-
- Carrasco, L., R. Guinea, A. Irurzun, and A. Barco. 2002. Effects of viral replication on cellular membrane metabolism and function, p. 337-354. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
-
- Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

