Evolutionary turnover of mammalian transcription start sites
- PMID: 16687732
- PMCID: PMC1473182
- DOI: 10.1101/gr.5031006
Evolutionary turnover of mammalian transcription start sites
Erratum in
- Genome Res. 2006 Jul;16(7):947. Hayshizaki, Yoshihide [corrected to Hayashizaki, Yoshihide]
Abstract
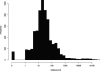
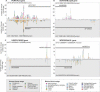
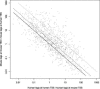
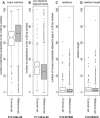
Alignments of homologous genomic sequences are widely used to identify functional genetic elements and study their evolution. Most studies tacitly equate homology of functional elements with sequence homology. This assumption is violated by the phenomenon of turnover, in which functionally equivalent elements reside at locations that are nonorthologous at the sequence level. Turnover has been demonstrated previously for transcription-factor-binding sites. Here, we show that transcription start sites of equivalent genes do not always reside at equivalent locations in the human and mouse genomes. We also identify two types of partial turnover, illustrating evolutionary pathways that could lead to complete turnover. These findings suggest that the signals encoding transcription start sites are highly flexible and evolvable, and have cautionary implications for the use of sequence-level conservation to detect gene regulatory elements.
Figures
Similar articles
-
Integrated analysis sheds light on evolutionary trajectories of young transcription start sites in the human genome.Genome Res. 2018 May;28(5):676-688. doi: 10.1101/gr.231449.117. Epub 2018 Apr 4. Genome Res. 2018. PMID: 29618487 Free PMC article.
-
Phylogenetic simulation of promoter evolution: estimation and modeling of binding site turnover events and assessment of their impact on alignment tools.Genome Biol. 2007;8(10):R225. doi: 10.1186/gb-2007-8-10-r225. Genome Biol. 2007. PMID: 17956628 Free PMC article.
-
SNP and indel frequencies at transcription start sites and at canonical and alternative translation initiation sites in the human genome.PLoS One. 2019 Apr 12;14(4):e0214816. doi: 10.1371/journal.pone.0214816. eCollection 2019. PLoS One. 2019. PMID: 30978217 Free PMC article.
-
Mammalian RNA polymerase II core promoters: insights from genome-wide studies.Nat Rev Genet. 2007 Jun;8(6):424-36. doi: 10.1038/nrg2026. Epub 2007 May 8. Nat Rev Genet. 2007. PMID: 17486122 Review.
-
Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex.Prog Nucleic Acid Res Mol Biol. 1998;61:309-44. doi: 10.1016/s0079-6603(08)60830-2. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752724 Review.
Cited by
-
Mapping of transcription start sites of human retina expressed genes.BMC Genomics. 2007 Feb 7;8:42. doi: 10.1186/1471-2164-8-42. BMC Genomics. 2007. PMID: 17286855 Free PMC article.
-
Systematic clustering of transcription start site landscapes.PLoS One. 2011;6(8):e23409. doi: 10.1371/journal.pone.0023409. Epub 2011 Aug 24. PLoS One. 2011. PMID: 21887249 Free PMC article.
-
28-way vertebrate alignment and conservation track in the UCSC Genome Browser.Genome Res. 2007 Dec;17(12):1797-808. doi: 10.1101/gr.6761107. Epub 2007 Nov 5. Genome Res. 2007. PMID: 17984227 Free PMC article.
-
The complexity of the mammalian transcriptome.J Physiol. 2006 Sep 1;575(Pt 2):321-32. doi: 10.1113/jphysiol.2006.115568. Epub 2006 Jul 20. J Physiol. 2006. PMID: 16857706 Free PMC article. Review.
-
The frequent evolutionary birth and death of functional promoters in mouse and human.Genome Res. 2015 Oct;25(10):1546-57. doi: 10.1101/gr.190546.115. Epub 2015 Jul 30. Genome Res. 2015. PMID: 26228054 Free PMC article.
References
-
- Anusaksathien O., Laplace C., Li X., Ren Y., Peng L., Goldring S.R., Galson D.L., Laplace C., Li X., Ren Y., Peng L., Goldring S.R., Galson D.L., Li X., Ren Y., Peng L., Goldring S.R., Galson D.L., Ren Y., Peng L., Goldring S.R., Galson D.L., Peng L., Goldring S.R., Galson D.L., Goldring S.R., Galson D.L., Galson D.L. Tissue-specific and ubiquitous promoters direct the expression of alternatively spliced transcripts from the calcitonin receptor gene. J. Biol. Chem. 2001;276:22663–22674. - PubMed
-
- Atanasova E., Chiappa S., Wieben E., Brimijoin S., Chiappa S., Wieben E., Brimijoin S., Wieben E., Brimijoin S., Brimijoin S. Novel messenger RNA and alternative promoter for murine acetylcholinesterase. J. Biol. Chem. 1999;274:21078–21084. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
