Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing
- PMID: 16682562
- PMCID: PMC1484422
- DOI: 10.1261/rna.30706
Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing
Abstract
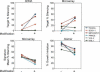
Transfected siRNAs regulate numerous transcripts sharing limited complementarity to the RNA duplex. This unintended ("off-target") silencing can hinder the use of RNAi to define gene function. Here we describe position-specific, sequence-independent chemical modifications that reduced silencing of partially complementary transcripts by all siRNAs tested. Silencing of perfectly matched targets was unaffected by these modifications. The chemical modification also reduced off-target phenotypes in growth inhibition studies. Key to the modification was 2'-O-methyl ribosyl substitution at position 2 in the guide strand, which reduced silencing of most off-target transcripts with complementarity to the seed region of the siRNA guide strand. The sharp position dependence of 2'-O-methyl ribosyl modification contrasts with the broader position dependence of base-pair substitutions within the seed region, suggesting a role for position 2 of the guide strand distinct from its effects on pairing to target transcripts.
Figures
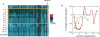


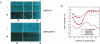

Similar articles
-
Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity.RNA. 2006 Jul;12(7):1179-87. doi: 10.1261/rna.25706. Epub 2006 May 8. RNA. 2006. PMID: 16682560 Free PMC article.
-
Modified siRNA structure with a single nucleotide bulge overcomes conventional siRNA-mediated off-target silencing.Mol Ther. 2011 Sep;19(9):1676-87. doi: 10.1038/mt.2011.109. Epub 2011 Jun 14. Mol Ther. 2011. PMID: 21673662 Free PMC article.
-
The siRNA Non-seed Region and Its Target Sequences Are Auxiliary Determinants of Off-Target Effects.PLoS Comput Biol. 2015 Dec 11;11(12):e1004656. doi: 10.1371/journal.pcbi.1004656. eCollection 2015 Dec. PLoS Comput Biol. 2015. PMID: 26657993 Free PMC article.
-
Is the Efficiency of RNA Silencing Evolutionarily Regulated?Int J Mol Sci. 2016 May 12;17(5):719. doi: 10.3390/ijms17050719. Int J Mol Sci. 2016. PMID: 27187367 Free PMC article. Review.
-
Chemical modification of siRNA.Curr Opin Mol Ther. 2010 Apr;12(2):158-67. Curr Opin Mol Ther. 2010. PMID: 20373259 Review.
Cited by
-
Specificity of oligonucleotide gene therapy (OGT) agents.Theranostics. 2022 Oct 9;12(16):7132-7157. doi: 10.7150/thno.77830. eCollection 2022. Theranostics. 2022. PMID: 36276652 Free PMC article. Review.
-
Development of modified siRNA molecules incorporating 5-fluoro-2'-deoxyuridine residues to enhance cytotoxicity.Nucleic Acids Res. 2013 Apr;41(8):4650-9. doi: 10.1093/nar/gkt120. Epub 2013 Feb 28. Nucleic Acids Res. 2013. PMID: 23449220 Free PMC article.
-
Milk Exosomes: Perspective Agents for Anticancer Drug Delivery.Int J Mol Sci. 2020 Sep 11;21(18):6646. doi: 10.3390/ijms21186646. Int J Mol Sci. 2020. PMID: 32932782 Free PMC article. Review.
-
siRNAs with decreased off-target effect facilitate the identification of essential genes in cancer cells.Oncotarget. 2015 Aug 28;6(25):21603-13. doi: 10.18632/oncotarget.4269. Oncotarget. 2015. PMID: 26057633 Free PMC article.
-
Thermodynamic Control of Small RNA-Mediated Gene Silencing.Front Genet. 2012 Jun 4;3:101. doi: 10.3389/fgene.2012.00101. eCollection 2012. Front Genet. 2012. PMID: 22675333 Free PMC article.
References
-
- Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. - PubMed
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Blake K., Murakami A., Miller P. Hybridization arrest of globin synthesis in rabbit reticulocyte lysates and cells by oligodeoxyribonucleoside methylphosphonates. Biochemistry. 1985;24:6139–6145. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
