E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment
- PMID: 16682500
- PMCID: PMC2121213
- DOI: 10.1084/jem.20060268
E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment
Abstract
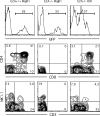
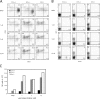
The helix-loop-helix protein, E47, is essential for both B- and T-lineage development. Here we demonstrate that in vitro E47 and Notch signaling act in concert to promote T cell development from fetal hematopoietic progenitors and to restrain development into the natural killer and myeloid cell lineages. The expression of an ensemble of genes associated with Notch signaling is activated by E47, and additionally, Notch signaling and E47 act in parallel pathways to induce a T lineage-specific program of gene expression. Enforced expression of the intracellular domain of Notch rescues the developmental arrest at the T cell commitment stage in E2A-deficient fetal thymocytes. Finally, we demonstrate that regulation of Hes1 expression by Notch signaling and E47 is strikingly similar to that observed during Drosophila melanogaster sensory development. Based on these observations, we propose that in developing fetal thymocytes E47 acts to induce the expression of an ensemble of genes involved in Notch signaling, and that subsequently E47 acts in parallel with Notch signaling to promote T-lineage maturation.
Figures

Similar articles
-
Context-dependent regulation of hematopoietic lineage choice by HEBAlt.J Immunol. 2010 Oct 1;185(7):4109-17. doi: 10.4049/jimmunol.0901783. Epub 2010 Sep 8. J Immunol. 2010. PMID: 20826759
-
Early thymocyte development is regulated by modulation of E2A protein activity.J Exp Med. 2001 Sep 17;194(6):733-45. doi: 10.1084/jem.194.6.733. J Exp Med. 2001. PMID: 11560990 Free PMC article.
-
Identification of Notch target genes in uncommitted T-cell progenitors: No direct induction of a T-cell specific gene program.Leukemia. 2006 Nov;20(11):1967-77. doi: 10.1038/sj.leu.2404396. Epub 2006 Sep 21. Leukemia. 2006. PMID: 16990763
-
Stepwise specification of lymphocyte developmental lineages.Curr Opin Genet Dev. 2000 Aug;10(4):370-9. doi: 10.1016/s0959-437x(00)00098-8. Curr Opin Genet Dev. 2000. PMID: 10889068 Review.
-
Regulation of lymphocyte development by Notch signaling.Nat Immunol. 2007 May;8(5):451-6. doi: 10.1038/ni1453. Nat Immunol. 2007. PMID: 17440450 Review.
Cited by
-
Launching the T-cell-lineage developmental programme.Nat Rev Immunol. 2008 Jan;8(1):9-21. doi: 10.1038/nri2232. Nat Rev Immunol. 2008. PMID: 18097446 Free PMC article. Review.
-
Helix-Loop-Helix Proteins in Adaptive Immune Development.Front Immunol. 2022 May 12;13:881656. doi: 10.3389/fimmu.2022.881656. eCollection 2022. Front Immunol. 2022. PMID: 35634342 Free PMC article. Review.
-
The perception and response of T cells to a changing environment are based on the law of initial value.Sci Signal. 2022 May 31;15(736):eabj9842. doi: 10.1126/scisignal.abj9842. Epub 2022 May 31. Sci Signal. 2022. PMID: 35639856 Free PMC article. Review.
-
Id1 attenuates Notch signaling and impairs T-cell commitment by elevating Deltex1 expression.Mol Cell Biol. 2009 Sep;29(17):4640-52. doi: 10.1128/MCB.00119-09. Epub 2009 Jun 29. Mol Cell Biol. 2009. PMID: 19564409 Free PMC article.
-
Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3.Nat Immunol. 2011 Jan;12(1):86-95. doi: 10.1038/ni.1965. Epub 2010 Dec 5. Nat Immunol. 2011. PMID: 21131965 Free PMC article.
References
-
- Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, G.F. Beilhack, J.A. Shimizu, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. - PubMed
-
- Busslinger, M. 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22:55–79. - PubMed
-
- Pelayo, R., R. Welner, S.S. Perry, J. Huang, Y. Baba, Y. Yokota, and P.W. Kincade. 2005. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr. Opin. Immunol. 17:100–107. - PubMed
-
- Katsura, Y. 2002. Redefinition of lymphoid progenitors. Nat. Rev. Immunol. 2:127–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

