Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus
- PMID: 16638566
- PMCID: PMC7094648
- DOI: 10.1016/j.febslet.2006.03.066
Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus
Abstract
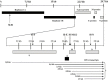
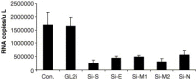
SARS-associated coronavirus was identified as the etiological agent of severe acute respiratory syndrome and a large virus pool was identified in wild animals. Virus generates drug resistance through fast mutagenesis and escapes antiviral treatment. siRNAs targeting different genes would be an alternative for overcoming drug resistance. Here, we report effective siRNAs targeting structural genes (i.e., spike, envelope, membrane, and nucleocapsid) and their antiviral kinetics. We also showed the synergistic effects of two siRNAs targeting different functional genes at a very low dose. Our findings may pave a way to develop cost effective siRNA agents for antiviral therapy in the future.
Figures


Similar articles
-
Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus.Antivir Ther. 2004 Jun;9(3):365-74. Antivir Ther. 2004. PMID: 15259899
-
siRNA targeting the leader sequence of SARS-CoV inhibits virus replication.Gene Ther. 2005 May;12(9):751-61. doi: 10.1038/sj.gt.3302479. Gene Ther. 2005. PMID: 15772689 Free PMC article.
-
Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins.J Virol. 2004 Sep;78(18):9977-86. doi: 10.1128/JVI.78.18.9977-9986.2004. J Virol. 2004. PMID: 15331731 Free PMC article.
-
Characterization of viral proteins encoded by the SARS-coronavirus genome.Antiviral Res. 2005 Feb;65(2):69-78. doi: 10.1016/j.antiviral.2004.10.001. Antiviral Res. 2005. PMID: 15708633 Free PMC article. Review.
-
Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus.Infect Disord Drug Targets. 2007 Mar;7(1):29-41. doi: 10.2174/187152607780090739. Infect Disord Drug Targets. 2007. PMID: 17346209 Review.
Cited by
-
Metabolic alterations upon SARS-CoV-2 infection and potential therapeutic targets against coronavirus infection.Signal Transduct Target Ther. 2023 Jun 7;8(1):237. doi: 10.1038/s41392-023-01510-8. Signal Transduct Target Ther. 2023. PMID: 37286535 Free PMC article. Review.
-
From a genome-wide screen of RNAi molecules against SARS-CoV-2 to a validated broad-spectrum and potent prophylaxis.Commun Biol. 2023 Mar 16;6(1):277. doi: 10.1038/s42003-023-04589-5. Commun Biol. 2023. PMID: 36928598 Free PMC article.
-
Genome wide screen of RNAi molecules against SARS-CoV-2 creates a broadly potent prophylaxis.bioRxiv [Preprint]. 2022 Apr 12:2022.04.12.488010. doi: 10.1101/2022.04.12.488010. bioRxiv. 2022. Update in: Commun Biol. 2023 Mar 16;6(1):277. doi: 10.1038/s42003-023-04589-5 PMID: 35441162 Free PMC article. Updated. Preprint.
-
Antisense technology as a potential strategy for the treatment of coronaviruses infection: With focus on COVID-19.IET Nanobiotechnol. 2022 May;16(3):67-77. doi: 10.1049/nbt2.12079. Epub 2022 Mar 10. IET Nanobiotechnol. 2022. PMID: 35274474 Free PMC article. Review.
-
Nucleic Acid-Based Treatments Against COVID-19: Potential Efficacy of Aptamers and siRNAs.Front Microbiol. 2021 Nov 8;12:758948. doi: 10.3389/fmicb.2021.758948. eCollection 2021. Front Microbiol. 2021. PMID: 34858370 Free PMC article. Review.
References
-
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med., 348, (2003), 1986– 1994. - PubMed
-
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med., 348, (2003), 1977– 1985. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

