Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein
- PMID: 16611922
- PMCID: PMC1472005
- DOI: 10.1128/JVI.80.9.4623-4632.2006
Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein
Abstract
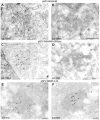
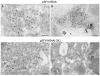
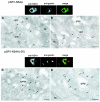
A common feature associated with the replication of most RNA viruses is the formation of a unique membrane environment encapsulating the viral replication complex. For their part, flaviviruses are no exception, whereupon infection causes a dramatic rearrangement and induction of unique membrane structures within the cytoplasm of infected cells. These virus-induced membranes, termed paracrystalline arrays, convoluted membranes, and vesicle packets, all appear to have specific functions during replication and are derived from different organelles within the host cell. The aim of this study was to identify which protein(s) specified by the Australian strain of West Nile virus, Kunjin virus (KUNV), are responsible for the dramatic membrane alterations observed during infection. Thus, we have shown using immunolabeling of ultrathin cryosections of transfected cells that expression of the KUNV polyprotein intermediates NS4A-4B and NS2B-3-4A, as well as that of individual NS4A proteins with and without the C-terminal transmembrane domain 2K, resulted in different degrees of rearrangement of cytoplasmic membranes. The formation of the membrane structures characteristic for virus infection required coexpression of an NS4A-NS4B cassette with the viral protease NS2B-3pro which was shown to be essential for the release of the individual NS4A and NS4B proteins. Individual expression of NS4A protein retaining the C-terminal transmembrane domain 2K resulted in the induction of membrane rearrangements most resembling virus-induced structures, while removal of the 2K domain led to a less profound membrane rearrangement but resulted in the redistribution of the NS4A protein to the Golgi apparatus. The results show that cleavage of the KUNV polyprotein NS4A-4B by the viral protease is the key initiation event in the induction of membrane rearrangement and that the NS4A protein intermediate containing the uncleaved C-terminal transmembrane domain plays an essential role in these membrane rearrangements.
Figures




Similar articles
-
The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner.J Biol Chem. 2007 Mar 23;282(12):8873-82. doi: 10.1074/jbc.M609919200. Epub 2007 Feb 2. J Biol Chem. 2007. PMID: 17276984
-
Determinants in Nonstructural Protein 4A of Dengue Virus Required for RNA Replication and Replication Organelle Biogenesis.J Virol. 2021 Oct 13;95(21):e0131021. doi: 10.1128/JVI.01310-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379504 Free PMC article.
-
Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site.J Virol. 1993 Apr;67(4):2327-35. doi: 10.1128/JVI.67.4.2327-2335.1993. J Virol. 1993. PMID: 8445732 Free PMC article.
-
Kunjin RNA replication and applications of Kunjin replicons.Adv Virus Res. 2003;59:99-140. doi: 10.1016/s0065-3527(03)59004-2. Adv Virus Res. 2003. PMID: 14696328 Review.
-
[Kunjin virus replicon--a novel viral vector].Sheng Wu Gong Cheng Xue Bao. 2011 Feb;27(2):141-6. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 21650037 Review. Chinese.
Cited by
-
Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation.Nat Commun. 2024 Jul 19;15(1):6080. doi: 10.1038/s41467-024-50437-3. Nat Commun. 2024. PMID: 39030239 Free PMC article.
-
Flavivirus Zika NS4A protein forms large oligomers in liposomes and in mild detergent.Sci Rep. 2024 May 31;14(1):12533. doi: 10.1038/s41598-024-63407-y. Sci Rep. 2024. PMID: 38822066 Free PMC article.
-
Classical swine fever virus non-structural protein 4A recruits dihydroorotate dehydrogenase to facilitate viral replication.J Virol. 2024 Jun 13;98(6):e0049424. doi: 10.1128/jvi.00494-24. Epub 2024 May 17. J Virol. 2024. PMID: 38757985
-
A Zika virus protein expression screen in Drosophila to investigate targeted host pathways during development.Dis Model Mech. 2024 Feb 1;17(2):dmm050297. doi: 10.1242/dmm.050297. Epub 2024 Feb 28. Dis Model Mech. 2024. PMID: 38214058 Free PMC article.
-
Autophagy of the ER: the secretome finds the lysosome.FEBS J. 2023 Dec;290(24):5656-5673. doi: 10.1111/febs.16986. Epub 2023 Nov 3. FEBS J. 2023. PMID: 37920925 Free PMC article. Review.
References
-
- Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. - PubMed
-
- Chu, P. W., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. - PubMed
-
- Chu, P. W., E. G. Westaway, and G. Coia. 1992. Comparison of centrifugation methods for molecular and morphological analysis of membranes associated with RNA replication of the flavivirus Kunjin. J. Virol. Methods 37:219-234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

