Expression profiles of endogenous retroviruses in Old World monkeys
- PMID: 16611901
- PMCID: PMC1472034
- DOI: 10.1128/JVI.80.9.4415-4421.2006
Expression profiles of endogenous retroviruses in Old World monkeys
Abstract
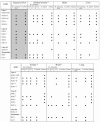
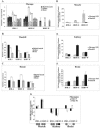
Human endogenous retroviruses (HERVs) are a major component of the human genome and an active part of the transcriptome. Some HERVs play vital biological roles, while others potentially contribute to diseases. Many HERVs are relatively new in the primate genome, having entered or expanded after the lineages leading to the platyrrhines (New World monkeys) and catarrhines (Old World monkeys and apes) separated. Most HERVs are active in at least some tissues, though tissue specificity is common for most elements. We analyzed multiple tissues from several Old World monkeys using retroviral pol-based DNA microarrays and quantitative PCR methods to determine their ERV expression profiles. The results demonstrate that while many ERVs are active in nonhuman primates, overall the tissue expression specificity is unique to each species. Most striking is that while the majority of HERVs analyzed in this study are expressed in human brain, almost none are expressed in Old World monkey brains or are only weakly expressed.
Figures
Similar articles
-
The distribution of pol containing human endogenous retroviruses in non-human primates.Virology. 2005 Apr 10;334(2):203-13. doi: 10.1016/j.virol.2005.01.045. Virology. 2005. PMID: 15780870
-
Origin and Deep Evolution of Human Endogenous Retroviruses in Pan-Primates.Viruses. 2022 Jun 23;14(7):1370. doi: 10.3390/v14071370. Viruses. 2022. PMID: 35891351 Free PMC article.
-
Human endogenous retroviruses: from infectious elements to human genes.Cytogenet Genome Res. 2005;110(1-4):318-32. doi: 10.1159/000084964. Cytogenet Genome Res. 2005. PMID: 16093684 Review.
-
Short communication: expression profiles of endogenous retroviral envelopes in Macaca mulatta (rhesus monkey).AIDS Res Hum Retroviruses. 2014 Oct;30(10):996-1000. doi: 10.1089/AID.2014.0010. Epub 2014 Jul 22. AIDS Res Hum Retroviruses. 2014. PMID: 24961963
-
Retroviruses and primate evolution.Bioessays. 2000 Feb;22(2):161-71. doi: 10.1002/(SICI)1521-1878(200002)22:2<161::AID-BIES7>3.0.CO;2-X. Bioessays. 2000. PMID: 10655035 Review.
Cited by
-
A novel function of RNAs arising from the long terminal repeat of human endogenous retrovirus 9 in cell cycle arrest.J Virol. 2013 Jan;87(1):25-36. doi: 10.1128/JVI.01648-12. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097441 Free PMC article.
-
Transmission, Evolution, and Endogenization: Lessons Learned from Recent Retroviral Invasions.Microbiol Mol Biol Rev. 2017 Dec 13;82(1):e00044-17. doi: 10.1128/MMBR.00044-17. Print 2018 Mar. Microbiol Mol Biol Rev. 2017. PMID: 29237726 Free PMC article. Review.
-
Transposable elements in TDP-43-mediated neurodegenerative disorders.PLoS One. 2012;7(9):e44099. doi: 10.1371/journal.pone.0044099. Epub 2012 Sep 5. PLoS One. 2012. PMID: 22957047 Free PMC article.
-
A novel endogenous betaretrovirus group characterized from polar bears (Ursus maritimus) and giant pandas (Ailuropoda melanoleuca).Virology. 2013 Aug 15;443(1):1-10. doi: 10.1016/j.virol.2013.05.008. Epub 2013 May 29. Virology. 2013. PMID: 23725819 Free PMC article.
-
Expression of evolutionarily novel genes in tumors.Infect Agent Cancer. 2016 Jul 19;11:34. doi: 10.1186/s13027-016-0077-6. eCollection 2016. Infect Agent Cancer. 2016. PMID: 27437030 Free PMC article. Review.
References
-
- Antony, J. M., G. van Marle, W. Opii, D. A. Butterfield, F. Mallet, V. W. Yong, J. L. Wallace, R. M. Deacon, K. Warren, and C. Power. 2004. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 7:1088-1095. - PubMed
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
-
- Belshaw, R., A. Katzourakis, J. Paces, A. Burt, and M. Tristem. 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22:814-817. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

