Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome
- PMID: 16606705
- PMCID: PMC1457046
- DOI: 10.1101/gr.4887606
Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome
Abstract
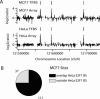
The E2F family of transcription factors regulates basic cellular processes. Here, we take an unbiased approach towards identifying E2F1 target genes by examining localization of E2F1-binding sites using high-density oligonucleotide tiling arrays. To begin, we developed a statistically-based methodology for analysis of ChIP-chip data obtained from arrays that represent 30 Mb of the human genome. Using this methodology, we identified regions bound by E2F1, MYC, and RNA Polymerase II (POLR2A). We found a large number of binding sites for all three factors; extrapolation suggests there may be approximately 20,000-30,000 E2F1- and MYC-binding sites and approximately 12,000-17,000 active promoters in HeLa cells. In contrast to our results for MYC, we find that the majority of E2F1-binding sites (>80%) are located in core promoters and that 50% of the sites overlap transcription starts. Only a small fraction of E2F1 sites possess the canonical binding motif. Surprisingly, we found that approximately 30% of genes in the 30-Mb region possessed an E2F1 binding site in a core promoter and E2F1 was bound near to 83% of POLR2A-bound sites. To determine if these results were representative of the entire human genome, we performed ChIP-chip analyses of approximately 24,000 promoters and confirmed that greater than 20% of the promoters were bound by E2F1. Our results suggest that E2F1 is recruited to promoters via a method distinct from recognition of the known consensus site and point toward a new understanding of E2F1 as a factor that contributes to the regulation of a large fraction of human genes.
Figures

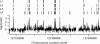

Similar articles
-
A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data--a case study using E2F1.Genome Res. 2006 Dec;16(12):1585-95. doi: 10.1101/gr.5520206. Epub 2006 Oct 19. Genome Res. 2006. PMID: 17053090 Free PMC article.
-
A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members.Genome Res. 2007 Nov;17(11):1550-61. doi: 10.1101/gr.6783507. Epub 2007 Oct 1. Genome Res. 2007. PMID: 17908821 Free PMC article.
-
E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites.Genome Res. 2008 Nov;18(11):1763-77. doi: 10.1101/gr.080622.108. Epub 2008 Oct 3. Genome Res. 2008. PMID: 18836037 Free PMC article.
-
Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome.J Biol Chem. 2011 Apr 8;286(14):11985-96. doi: 10.1074/jbc.M110.217158. Epub 2011 Feb 10. J Biol Chem. 2011. PMID: 21310950 Free PMC article.
-
Transcriptional regulation of the mouse PNRC2 promoter by the nuclear factor Y (NFY) and E2F1.Gene. 2005 Nov 21;361:89-100. doi: 10.1016/j.gene.2005.07.012. Epub 2005 Sep 21. Gene. 2005. PMID: 16181749
Cited by
-
MOST+: A de novo motif finding approach combining genomic sequence and heterogeneous genome-wide signatures.BMC Genomics. 2015;16 Suppl 7(Suppl 7):S13. doi: 10.1186/1471-2164-16-S7-S13. Epub 2015 Jun 11. BMC Genomics. 2015. PMID: 26099518 Free PMC article.
-
Uncovering transcription factor modules using one- and three-dimensional analyses.J Biol Chem. 2012 Sep 7;287(37):30914-21. doi: 10.1074/jbc.R111.309229. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952238 Free PMC article. Review.
-
Suz12 binds to silenced regions of the genome in a cell-type-specific manner.Genome Res. 2006 Jul;16(7):890-900. doi: 10.1101/gr.5306606. Epub 2006 Jun 2. Genome Res. 2006. PMID: 16751344 Free PMC article.
-
Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5.Mol Cell Biol. 2007 Oct;27(19):6889-902. doi: 10.1128/MCB.00762-07. Epub 2007 Jul 16. Mol Cell Biol. 2007. PMID: 17636019 Free PMC article.
-
The transcription unit architecture of the Escherichia coli genome.Nat Biotechnol. 2009 Nov;27(11):1043-9. doi: 10.1038/nbt.1582. Epub 2009 Nov 1. Nat Biotechnol. 2009. PMID: 19881496 Free PMC article.
References
-
- Brehm A., Miska E.A., McCance D.J., Reid J.L., Bannister A.J., Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
