Esc4/Rtt107 and the control of recombination during replication
- PMID: 16569515
- PMCID: PMC2881479
- DOI: 10.1016/j.dnarep.2006.02.005
Esc4/Rtt107 and the control of recombination during replication
Abstract
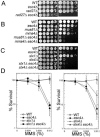
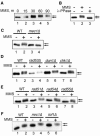
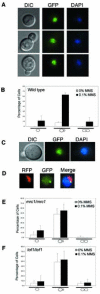
When replication forks stall during DNA synthesis, cells respond by assembling multi-protein complexes to control the various pathways that stabilize the replication machinery, repair the replication fork, and facilitate the reinitiation of processive DNA synthesis. Increasing evidence suggests that cells have evolved scaffolding proteins to orchestrate and control the assembly of these repair complexes, typified in mammalian cells by several BRCT-motif containing proteins, such as Brca1, Xrcc1, and 53BP1. In Saccharomyces cerevisiae, Esc4 contains six such BRCT domains and is required for the most efficient response to a variety of agents that damage DNA. We show that Esc4 interacts with several proteins involved in the repair and processing of stalled or collapsed replication forks, including the recombination protein Rad55. However, the function of Esc4 does not appear to be restricted to a Rad55-dependent process, as we observed an increase in sensitivity to the DNA alkylating agent methane methylsulfonate (MMS) in a esc4Deltarad55Delta mutant, as well as in double mutants of esc4Delta and other recombination genes, compared to the corresponding single mutants. In addition, we show that Esc4 forms multiple nuclear foci in response to treatment with MMS. Similar behavior is also observed in the absence of damage when either of the S-phase checkpoint proteins, Tof1 or Mrc1, is deleted. Thus, we propose that Esc4 associates with ssDNA of stalled forks and acts as a scaffolding protein to recruit and/or modulate the function of other proteins required to reinitiate DNA synthesis.
Figures




Similar articles
-
Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks.Mol Cell Biol. 2006 Nov;26(22):8396-409. doi: 10.1128/MCB.01317-06. Epub 2006 Sep 11. Mol Cell Biol. 2006. PMID: 16966380 Free PMC article.
-
Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs.BMC Mol Biol. 2006 Nov 9;7:40. doi: 10.1186/1471-2199-7-40. BMC Mol Biol. 2006. PMID: 17094803 Free PMC article.
-
Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae.DNA Repair (Amst). 2010 Oct 5;9(10):1038-49. doi: 10.1016/j.dnarep.2010.07.003. Epub 2010 Jul 31. DNA Repair (Amst). 2010. PMID: 20674515
-
DNA structure dependent checkpoints as regulators of DNA repair.DNA Repair (Amst). 2002 Dec 5;1(12):983-94. doi: 10.1016/s1568-7864(02)00165-9. DNA Repair (Amst). 2002. PMID: 12531008 Review.
-
ATR/Mec1: coordinating fork stability and repair.Curr Opin Cell Biol. 2009 Apr;21(2):237-44. doi: 10.1016/j.ceb.2009.01.017. Epub 2009 Feb 21. Curr Opin Cell Biol. 2009. PMID: 19230642 Review.
Cited by
-
Multi-BRCT scaffolds use distinct strategies to support genome maintenance.Cell Cycle. 2016 Oct;15(19):2561-2570. doi: 10.1080/15384101.2016.1218102. Epub 2016 Aug 11. Cell Cycle. 2016. PMID: 27580271 Free PMC article. Review.
-
Conditional genetic interactions of RTT107, SLX4, and HRQ1 reveal dynamic networks upon DNA damage in S. cerevisiae.G3 (Bethesda). 2014 Apr 2;4(6):1059-69. doi: 10.1534/g3.114.011205. G3 (Bethesda). 2014. PMID: 24700328 Free PMC article.
-
Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase.EMBO J. 2009 Apr 22;28(8):1121-30. doi: 10.1038/emboj.2009.43. Epub 2009 Mar 5. EMBO J. 2009. PMID: 19262568 Free PMC article.
-
gammaH2A binds Brc1 to maintain genome integrity during S-phase.EMBO J. 2010 Mar 17;29(6):1136-48. doi: 10.1038/emboj.2009.413. Epub 2010 Jan 21. EMBO J. 2010. PMID: 20094029 Free PMC article.
-
Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases.Genes (Basel). 2020 Feb 18;11(2):205. doi: 10.3390/genes11020205. Genes (Basel). 2020. PMID: 32085395 Free PMC article. Review.
References
-
- Tercero JA, Diffley JFX. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. - PubMed
-
- Santocanale C, Diffley JFX. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. - PubMed
-
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

