Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3
- PMID: 16567630
- PMCID: PMC1459354
- DOI: 10.1073/pnas.0510506103
Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3
Abstract
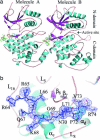
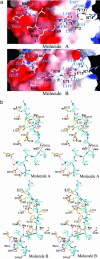
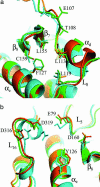
Mitogen-activated protein (MAP) kinases are central components of signal transduction pathways for cell proliferation, stress responses, and differentiation. Signaling efficiency and specificity are modulated in large part by docking interactions between individual MAP kinase and the kinase interaction motif (KIM), (R/K)(2-3)-X(1-6)-Phi(A)-X-Phi(B), in its cognate kinases, phosphatases, scaffolding proteins, and substrates. We have determined the crystal structure of extracellular signal-regulated protein kinase 2 bound to the KIM peptide from MAP kinase phosphatase 3, an extracellular signal-regulated protein kinase 2-specific phosphatase. The structure reveals that the KIM docking site, situated in a noncatalytic region opposite of the kinase catalytic pocket, is comprised of a highly acidic patch and a hydrophobic groove, which engage the basic and Phi(A)-X-Phi(B) residues, respectively, in the KIM sequence. The specific docking interactions observed in the structure consolidate all known biochemical data. In addition, structural comparison indicates that the KIM docking site is conserved in all MAP kinases. The results establish a structural model for understanding how MAP kinases interact with their regulators and substrates and provide new insights into how MAP kinase docking specificity can be achieved.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
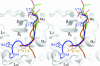
Similar articles
-
A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates.J Biol Chem. 2003 Aug 8;278(32):29901-12. doi: 10.1074/jbc.M303909200. Epub 2003 May 16. J Biol Chem. 2003. PMID: 12754209
-
Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation.J Mol Biol. 2001 Aug 17;311(3):557-68. doi: 10.1006/jmbi.2001.4890. J Mol Biol. 2001. PMID: 11493009
-
Docking interactions induce exposure of activation loop in the MAP kinase ERK2.Structure. 2006 Jun;14(6):1011-9. doi: 10.1016/j.str.2006.04.006. Structure. 2006. PMID: 16765894
-
Three-dimensional docking in the MAPK p38α.Sci Signal. 2011 Dec 20;4(204):pe47. doi: 10.1126/scisignal.2002697. Sci Signal. 2011. PMID: 22375047 Review.
-
Molecular basis of MAP kinase regulation.Protein Sci. 2013 Dec;22(12):1698-710. doi: 10.1002/pro.2374. Epub 2013 Oct 19. Protein Sci. 2013. PMID: 24115095 Free PMC article. Review.
Cited by
-
Sequence alignment reveals possible MAPK docking motifs on HIV proteins.PLoS One. 2010 Jan 28;5(1):e8942. doi: 10.1371/journal.pone.0008942. PLoS One. 2010. PMID: 20126615 Free PMC article.
-
Structural basis of p38α regulation by hematopoietic tyrosine phosphatase.Nat Chem Biol. 2011 Nov 6;7(12):916-24. doi: 10.1038/nchembio.707. Nat Chem Biol. 2011. PMID: 22057126 Free PMC article.
-
Structure of ERK2 bound to PEA-15 reveals a mechanism for rapid release of activated MAPK.Nat Commun. 2013;4:1681. doi: 10.1038/ncomms2687. Nat Commun. 2013. PMID: 23575685 Free PMC article.
-
ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4.Nat Struct Mol Biol. 2012 Feb 5;19(3):283-90. doi: 10.1038/nsmb.2217. Nat Struct Mol Biol. 2012. PMID: 22307056
-
MAPK/ERK Pathway as a Central Regulator in Vertebrate Organ Regeneration.Int J Mol Sci. 2022 Jan 27;23(3):1464. doi: 10.3390/ijms23031464. Int J Mol Sci. 2022. PMID: 35163418 Free PMC article. Review.
References
-
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L. Physiol. Rev. 1999;79:143–180. - PubMed
-
- Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H. Chem. Rev. 2001;101:2449–2476. - PubMed
-
- Holland P. M., Cooper J. A. Curr. Biol. 1999;9:R329–R331. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

