A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana
- PMID: 16567499
- PMCID: PMC2063757
- DOI: 10.1083/jcb.200508116
A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana
Abstract
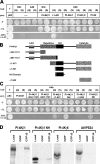
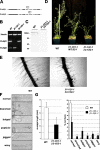
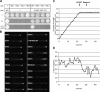
The RabA4b GTPase labels a novel, trans-Golgi network compartment displaying a developmentally regulated polar distribution in growing Arabidopsis thaliana root hair cells. GTP bound RabA4b selectively recruits the plant phosphatidylinositol 4-OH kinase, PI-4Kbeta1, but not members of other PI-4K families. PI-4Kbeta1 colocalizes with RabA4b on tip-localized membranes in growing root hairs, and mutant plants in which both the PI-4Kbeta1 and -4Kbeta2 genes are disrupted display aberrant root hair morphologies. PI-4Kbeta1 interacts with RabA4b through a novel homology domain, specific to eukaryotic type IIIbeta PI-4Ks, and PI-4Kbeta1 also interacts with a Ca2+ sensor, AtCBL1, through its NH2 terminus. We propose that RabA4b recruitment of PI-4Kbeta1 results in Ca2+-dependent generation of PI-4P on this compartment, providing a link between Ca2+ and PI-4,5P2-dependent signals during the polarized secretion of cell wall components in tip-growing root hair cells.
Figures


Similar articles
-
Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis.Traffic. 2011 Mar;12(3):313-29. doi: 10.1111/j.1600-0854.2010.01146.x. Epub 2011 Jan 7. Traffic. 2011. PMID: 21134079
-
The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells.Plant Cell. 2004 Jun;16(6):1589-603. doi: 10.1105/tpc.021634. Epub 2004 May 21. Plant Cell. 2004. PMID: 15155878 Free PMC article.
-
Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana.Plant Cell. 2008 Feb;20(2):381-95. doi: 10.1105/tpc.107.054304. Epub 2008 Feb 15. Plant Cell. 2008. PMID: 18281508 Free PMC article.
-
Emerging aspects of ER organization in root hair tip growth: lessons from RHD3 and Atlastin.Plant Signal Behav. 2011 Nov;6(11):1710-3. doi: 10.4161/psb.6.11.17477. Epub 2011 Nov 1. Plant Signal Behav. 2011. PMID: 22057320 Free PMC article. Review.
-
Building a hair: tip growth in Arabidopsis thaliana root hairs.Philos Trans R Soc Lond B Biol Sci. 2002 Jun 29;357(1422):815-21. doi: 10.1098/rstb.2002.1092. Philos Trans R Soc Lond B Biol Sci. 2002. PMID: 12079677 Free PMC article. Review.
Cited by
-
A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development.PLoS One. 2013;8(2):e56412. doi: 10.1371/journal.pone.0056412. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437128 Free PMC article.
-
Signalling Pinpointed to the Tip: The Complex Regulatory Network That Allows Pollen Tube Growth.Plants (Basel). 2020 Aug 26;9(9):1098. doi: 10.3390/plants9091098. Plants (Basel). 2020. PMID: 32859043 Free PMC article. Review.
-
Fine Structural Analysis of Brefeldin A-Induced Compartment Formation After High-Pressure Freeze Fixation of Maize Root Epidermis: Compound Exocytosis Resembling Cell Plate Formation during Cytokinesis.Plant Signal Behav. 2006 May;1(3):134-9. doi: 10.4161/psb.1.3.2996. Plant Signal Behav. 2006. PMID: 19521493 Free PMC article.
-
Subcellular localization and functional analysis of the Arabidopsis GTPase RabE.Plant Physiol. 2009 Apr;149(4):1824-37. doi: 10.1104/pp.108.132092. Epub 2009 Feb 20. Plant Physiol. 2009. PMID: 19233904 Free PMC article.
-
Unique mechanism of plant endocytic/vacuolar transport pathways.J Plant Res. 2009 Jan;122(1):21-30. doi: 10.1007/s10265-008-0200-x. Epub 2008 Dec 11. J Plant Res. 2009. PMID: 19082690
References
-
- Balla, T. 1998. Phosphatidylinositol 4-kinases. Biochim. Biophys. Acta. 1436:69–85. - PubMed
-
- Balla, A., G. Tuymetova, A. Tsiomenko, P. Varnai, and T. Balla. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 16:1282–1295. - PMC - PubMed
-
- Bankaitis, V.A., and A.J. Morris. 2003. Lipids and the exocytotic machinery of eukaryotic cells. Curr. Opin. Cell Biol. 15:389–395. - PubMed
-
- Bubb, M.R., I.C. Baines, and E.D. Korn. 1998. Localization of actobindin, profilin I, profilin II, and phosphatidylinositol-4,5-bisphosphate (PIP2) in Acanthamoeba castellanii. Cell Motil. Cytoskeleton. 39:134–146. - PubMed
-
- Christoforidis, S., and M. Zerial. 2000. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 20:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

