Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis
- PMID: 16549795
- PMCID: PMC1458802
- DOI: 10.1073/pnas.0508200103
Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis
Abstract
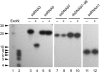
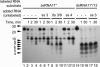
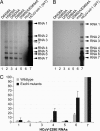
Replication of the giant RNA genome of severe acute respiratory syndrome (SARS) coronavirus (CoV) and synthesis of as many as eight subgenomic (sg) mRNAs are mediated by a viral replicase-transcriptase of outstanding complexity that includes an essential endoribonuclease activity. Here, we show that the CoV replicative machinery, unlike that of other RNA viruses, also uses an exoribonuclease (ExoN) activity, which is associated with nonstructural protein (nsp) 14. Bacterially expressed forms of SARS-CoV nsp14 were shown to act on both ssRNAs and dsRNAs in a 3'-->5' direction. The activity depended on residues that are conserved in the DEDD exonuclease superfamily. The protein did not hydrolyze DNA or ribose-2'-O-methylated RNA substrates and required divalent metal ions for activity. A range of 5'-labeled ssRNA substrates were processed to final products of approximately 8-12 nucleotides. When part of dsRNA or in the presence of nonlabeled dsRNA, the 5'-labeled RNA substrates were processed to significantly smaller products, indicating that binding to dsRNA in cis or trans modulates the exonucleolytic activity of nsp14. Characterization of human CoV 229E ExoN active-site mutants revealed severe defects in viral RNA synthesis, and no viable virus could be recovered. Besides strongly reduced genome replication, specific defects in sg RNA synthesis, such as aberrant sizes of specific sg RNAs and changes in the molar ratios between individual sg RNA species, were observed. Taken together, the study identifies an RNA virus ExoN activity that is involved in the synthesis of multiple RNAs from the exceptionally large genomic RNA templates of CoVs.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

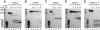
Similar articles
-
High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants.J Virol. 2007 Nov;81(22):12135-44. doi: 10.1128/JVI.01296-07. Epub 2007 Sep 5. J Virol. 2007. PMID: 17804504 Free PMC article.
-
Proofreading-Deficient Coronaviruses Adapt for Increased Fitness over Long-Term Passage without Reversion of Exoribonuclease-Inactivating Mutations.mBio. 2017 Nov 7;8(6):e01503-17. doi: 10.1128/mBio.01503-17. mBio. 2017. PMID: 29114026 Free PMC article.
-
The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2.J Virol. 2020 Nov 9;94(23):e01246-20. doi: 10.1128/JVI.01246-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938769 Free PMC article.
-
Biochemical aspects of coronavirus replication and virus-host interaction.Annu Rev Microbiol. 2006;60:211-30. doi: 10.1146/annurev.micro.60.080805.142157. Annu Rev Microbiol. 2006. PMID: 16712436 Review.
-
The structure and functions of coronavirus genomic 3' and 5' ends.Virus Res. 2015 Aug 3;206:120-33. doi: 10.1016/j.virusres.2015.02.025. Epub 2015 Feb 28. Virus Res. 2015. PMID: 25736566 Free PMC article. Review.
Cited by
-
Benefits of Repeated SARS-CoV-2 Vaccination and Virus-induced Cross-neutralization Potential in Immunocompromised Transplant Patients and Healthy Individuals.Open Forum Infect Dis. 2024 Sep 9;11(10):ofae527. doi: 10.1093/ofid/ofae527. eCollection 2024 Oct. Open Forum Infect Dis. 2024. PMID: 39371367 Free PMC article.
-
COVID-19 mutations: An overview.World J Methodol. 2024 Sep 20;14(3):89761. doi: 10.5662/wjm.v14.i3.89761. eCollection 2024 Sep 20. World J Methodol. 2024. PMID: 39310238 Free PMC article. Review.
-
Unchecked growth: Pushing the limits on RNA virus genome size in the absence of known proofreading.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2414223121. doi: 10.1073/pnas.2414223121. Epub 2024 Aug 26. Proc Natl Acad Sci U S A. 2024. PMID: 39186661 Free PMC article. No abstract available.
-
In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.Sci Adv. 2024 Jul 26;10(30):eadl4013. doi: 10.1126/sciadv.adl4013. Epub 2024 Jul 24. Sci Adv. 2024. PMID: 39047088 Free PMC article.
-
A ~40-kb flavi-like virus does not encode a known error-correcting mechanism.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2403805121. doi: 10.1073/pnas.2403805121. Epub 2024 Jul 17. Proc Natl Acad Sci U S A. 2024. PMID: 39018195 Free PMC article.
References
-
- Peiris J. S., Yuen K. Y., Osterhaus A. D., Stohr K. N. Engl. J. Med. 2003;349:2431–2441. - PubMed
-
- Siddell S. G., Ziebuhr J., Snijder E. J. In: Topley and Wilson's Microbiology and Microbial Infections. Mahy B. W. J., ter Meulen V., editors. London: Hodder Arnold; 2005. pp. 823–856.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

