Deciphering cellular states of innate tumor drug responses
- PMID: 16542501
- PMCID: PMC1557757
- DOI: 10.1186/gb-2006-7-3-r19
Deciphering cellular states of innate tumor drug responses
Abstract
Background: The molecular mechanisms underlying innate tumor drug resistance, a major obstacle to successful cancer therapy, remain poorly understood. In colorectal cancer (CRC), molecular studies have focused on drug-selected tumor cell lines or individual candidate genes using samples derived from patients already treated with drugs, so that very little data are available prior to drug treatment.
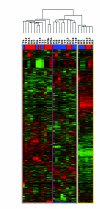
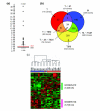
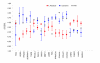
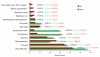
Results: Transcriptional profiles of clinical samples collected from CRC patients prior to their exposure to a combined chemotherapy of folinic acid, 5-fluorouracil and irinotecan were established using microarrays. Vigilant experimental design, power simulations and robust statistics were used to restrain the rates of false negative and false positive hybridizations, allowing successful discrimination between drug resistance and sensitivity states with restricted sampling. A list of 679 genes was established that intrinsically differentiates, for the first time prior to drug exposure, subsequently diagnosed chemo-sensitive and resistant patients. Independent biological validation performed through quantitative PCR confirmed the expression pattern on two additional patients. Careful annotation of interconnected functional networks provided a unique representation of the cellular states underlying drug responses.
Conclusion: Molecular interaction networks are described that provide a solid foundation on which to anchor working hypotheses about mechanisms underlying in vivo innate tumor drug responses. These broad-spectrum cellular signatures represent a starting point from which by-pass chemotherapy schemes, targeting simultaneously several of the molecular mechanisms involved, may be developed for critical therapeutic intervention in CRC patients. The demonstrated power of this research strategy makes it generally applicable to other physiological and pathological situations.
Figures

Similar articles
-
Establishment and characterization of cell lines from chromosomal instable colorectal cancer.World J Gastroenterol. 2015 Jan 7;21(1):164-76. doi: 10.3748/wjg.v21.i1.164. World J Gastroenterol. 2015. PMID: 25574089 Free PMC article.
-
APRIL is a novel clinical chemo-resistance biomarker in colorectal adenocarcinoma identified by gene expression profiling.BMC Cancer. 2009 Dec 11;9:434. doi: 10.1186/1471-2407-9-434. BMC Cancer. 2009. PMID: 20003335 Free PMC article.
-
Identification of candidate genes and miRNAs for sensitizing resistant colorectal cancer cells to oxaliplatin and irinotecan.Cancer Chemother Pharmacol. 2020 Jan;85(1):153-171. doi: 10.1007/s00280-019-03975-3. Epub 2019 Nov 28. Cancer Chemother Pharmacol. 2020. PMID: 31781855
-
[A review of chemotherapy for metastatic colon cancer].Farm Hosp. 2006 Nov-Dec;30(6):359-69. doi: 10.1016/s1130-6343(06)74007-7. Farm Hosp. 2006. PMID: 17298193 Review. Spanish.
-
DNA copy number profiles correlate with outcome in colorectal cancer patients treated with fluoropyrimidine/antifolate-based regimens.Curr Drug Metab. 2011 Dec;12(10):956-65. doi: 10.2174/138920011798062337. Curr Drug Metab. 2011. PMID: 21787269 Review.
Cited by
-
A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor.EMBO J. 2008 May 7;27(9):1368-77. doi: 10.1038/emboj.2008.61. Epub 2008 Apr 3. EMBO J. 2008. PMID: 18388863 Free PMC article.
-
Gene expression profiling and real-time PCR analyses identify novel potential cancer-testis antigens in multiple myeloma.J Immunol. 2009 Jul 15;183(2):832-40. doi: 10.4049/jimmunol.0803298. Epub 2009 Jun 19. J Immunol. 2009. PMID: 19542363 Free PMC article.
-
Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment.Cancer Cell. 2009 Jun 2;15(6):527-38. doi: 10.1016/j.ccr.2009.04.010. Cancer Cell. 2009. PMID: 19477431 Free PMC article.
-
Characterisation and correction of signal fluctuations in successive acquisitions of microarray images.BMC Bioinformatics. 2009 Mar 30;10:98. doi: 10.1186/1471-2105-10-98. BMC Bioinformatics. 2009. PMID: 19331668 Free PMC article.
-
INHBA is a novel mediator regulating cellular senescence and immune evasion in colorectal cancer.J Cancer. 2021 Aug 13;12(19):5938-5949. doi: 10.7150/jca.61556. eCollection 2021. J Cancer. 2021. PMID: 34476008 Free PMC article.
References
-
- Anonymous Modulation of fluorouracil by leucovorin in patients with advanced colorectal cancer: evidence in terms of response rate. Advanced Colorectal Cancer Meta-Analysis Project. J Clin Oncol. 1992;10:896–903. - PubMed
-
- Ychou M, Duffour J, Kramar A, Debrigode C, Gourgou S, Bressolle F, Pinguet F. Individual 5-FU dose adaptation in metastatic colorectal cancer: results of a phase II study using a bimonthly pharmacokinetically intensified LV5FU2 regimen. Cancer Chemother Pharmacol. 2003;52:282–290. doi: 10.1007/s00280-003-0658-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

